3ID3
 
 | | Crystal Structure of RseP PDZ2 I304A domain | | Descriptor: | Regulator of sigma E protease | | Authors: | Li, X, Wang, B, Feng, L, Wang, J, Shi, Y. | | Deposit date: | 2009-07-20 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Cleavage of RseA by RseP requires a carboxyl-terminal hydrophobic amino acid following DegS cleavage
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3Q8T
 
 | |
2GNC
 
 | | Crystal structure of srGAP1 SH3 domain in the slit-robo signaling pathway | | Descriptor: | SLIT-ROBO Rho GTPase-activating protein 1 | | Authors: | Li, X, Liu, Y, Gao, F, Bartlam, M, Wu, J.Y, Rao, Z. | | Deposit date: | 2006-04-10 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Robo Proline-rich Motif Recognition by the srGAP1 Src Homology 3 Domain in the Slit-Robo Signaling Pathway
J.Biol.Chem., 281, 2006
|
|
3RQF
 
 | |
3RQG
 
 | |
3RQE
 
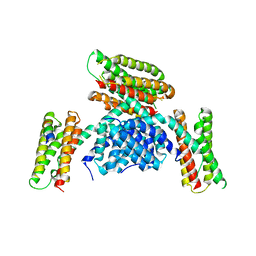 | |
2DC2
 
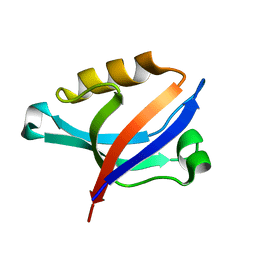 | | Solution Structure of PDZ Domain | | Descriptor: | golgi associated PDZ and coiled-coil motif containing isoform b | | Authors: | Li, X, Wu, J, Shi, Y. | | Deposit date: | 2005-12-20 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of GOPC PDZ domain and its interaction with the C-terminal motif of neuroligin
Protein Sci., 15, 2006
|
|
2HNU
 
 | | Crystal Structure of a Dipeptide Complex of Bovine Neurophysin-I | | Descriptor: | Oxytocin-neurophysin 1, PHENYLALANINE, TYROSINE | | Authors: | Li, X, Lee, H, Wu, J, Breslow, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contributions of the interdomain loop, amino terminus, and subunit interface to the ligand-facilitated dimerization of neurophysin: crystal structures and mutation studies of bovine neurophysin-I.
Protein Sci., 16, 2007
|
|
2HNV
 
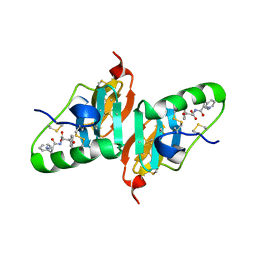 | | Crystal Structure of a Dipeptide Complex of the Q58V Mutant of Bovine Neurophysin-I | | Descriptor: | Oxytocin-neurophysin 1, PHENYLALANINE, TYROSINE | | Authors: | Li, X, Lee, H, Wu, J, Breslow, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contributions of the interdomain loop, amino terminus, and subunit interface to the ligand-facilitated dimerization of neurophysin: crystal structures and mutation studies of bovine neurophysin-I.
Protein Sci., 16, 2007
|
|
2HNW
 
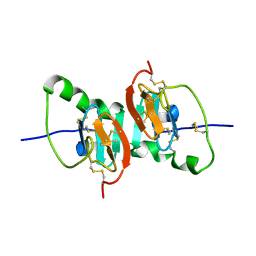 | | Crystal Structure of the F91STOP mutant of des1-6 Bovine Neurophysin-I, unliganded state | | Descriptor: | Oxytocin-neurophysin 1 | | Authors: | Li, X, Lee, H, Wu, J, Breslow, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-04-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Contributions of the interdomain loop, amino terminus, and subunit interface to the ligand-facilitated dimerization of neurophysin: crystal structures and mutation studies of bovine neurophysin-I.
Protein Sci., 16, 2007
|
|
4EIS
 
 | | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases (PMO-3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, PEROXIDE ION, ... | | Authors: | Li, X, Beeson, W.T, Phillips, C.M, Marletta, M.A, Cate, J.H. | | Deposit date: | 2012-04-05 | | Release date: | 2012-05-23 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases.
Structure, 20, 2012
|
|
4DXA
 
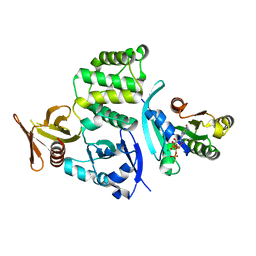 | | Co-crystal structure of Rap1 in complex with KRIT1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Krev interaction trapped protein 1, MAGNESIUM ION, ... | | Authors: | Li, X, Zhang, R, Boggon, T.J. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Small G Protein Effector Interaction of Ras-related Protein 1 (Rap1) and Adaptor Protein Krev Interaction Trapped 1 (KRIT1).
J.Biol.Chem., 287, 2012
|
|
4FWI
 
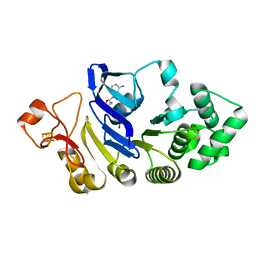 | | Crystal structure of the nucleotide-binding domain of a dipeptide ABC transporter | | Descriptor: | ABC-type dipeptide/oligopeptide/nickel transport system, ATPase component, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Li, X, Ge, J, Yang, M, Wang, N. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | Structure of the nucleotide-binding domain of a dipeptide ABC transporter reveals a novel iron-sulfur cluster-binding domain
Acta Crystallogr.,Sect.D, 69, 2013
|
|
9F3E
 
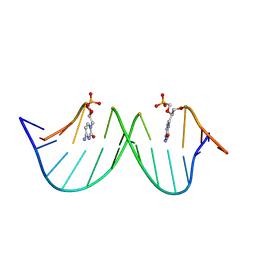 | |
4EIR
 
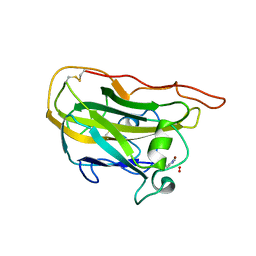 | | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases (PMO-2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | Authors: | Li, X, Beeson, W.T, Phillips, C.M, Marletta, M.A, Cate, J.H. | | Deposit date: | 2012-04-05 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases.
Structure, 20, 2012
|
|
3L8J
 
 | | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity | | Descriptor: | Programmed cell death protein 10 | | Authors: | Li, X, Zhang, R, Zhang, H, He, Y, Ji, W, Min, W, Boggon, T.J. | | Deposit date: | 2009-12-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity.
J.Biol.Chem., 285, 2010
|
|
3L8I
 
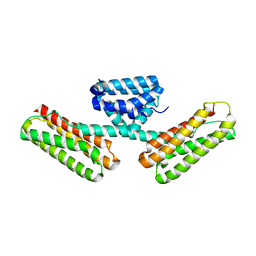 | | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity | | Descriptor: | Programmed cell death protein 10 | | Authors: | Li, X, Zhang, R, Zhang, H, He, Y, Ji, W, Min, W, Boggon, T.J. | | Deposit date: | 2009-12-31 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity.
J.Biol.Chem., 285, 2010
|
|
7N6R
 
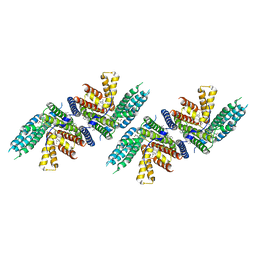 | | Structure of nevanimibe bound human ACAT2 | | Descriptor: | CHOLESTEROL, OLEIC ACID, Sterol O-acyltransferase 2, ... | | Authors: | Li, X, Long, T. | | Deposit date: | 2021-06-08 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structure of nevanimibe bound human ACAT2
To Be Published
|
|
7N6Q
 
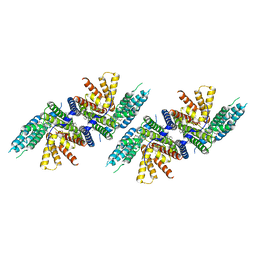 | | Structure of PPPA bound human ACAT2 | | Descriptor: | (3S,4R,4aR,6S,6aS,12R,12aS,12bS)-4-[(acetyloxy)methyl]-12-hydroxy-4,6a,12b-trimethyl-11-oxo-9-(pyridin-3-yl)-1,3,4,4a,5,6,6a,12,12a,12b-decahydro-2H,11H-naphtho[2,1-b]pyrano[3,4-e]pyran-3,6-diyl diacetate, CHOLESTEROL, OLEIC ACID, ... | | Authors: | Li, X, Long, T. | | Deposit date: | 2021-06-08 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structure of PPPA bound human ACAT2
To Be Published
|
|
5IND
 
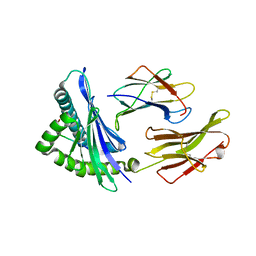 | | Crystal structure of HLA-B5801, a protective HLA allele for HIV-1 infection | | Descriptor: | Beta-2-microglobulin, GLN-ALA-SER-GLN-ASP-VAL-LYS-ASN-TRP, HLA class I histocompatibility antigen, ... | | Authors: | Li, X, Wang, J.-H. | | Deposit date: | 2016-03-07 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.132 Å) | | Cite: | Crystal structure of HLA-B*5801, a protective HLA allele for HIV-1 infection.
Protein Cell, 7, 2016
|
|
5INC
 
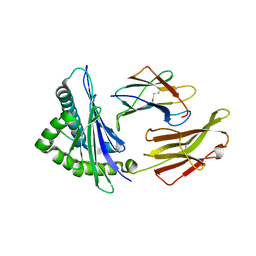 | | Crystal structure of HLA-B5801, a protective HLA allele for HIV-1 infection | | Descriptor: | Beta-2-microglobulin, GLN-ALA-THR-GLN-GLU-VAL-LYS-ASN-TRP, HLA class I histocompatibility antigen, ... | | Authors: | Li, X, Wang, J.-H. | | Deposit date: | 2016-03-07 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.881 Å) | | Cite: | Crystal structure of HLA-B*5801, a protective HLA allele for HIV-1 infection.
Protein Cell, 7, 2016
|
|
5IM7
 
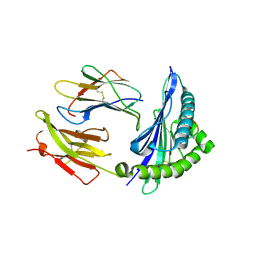 | |
7YMN
 
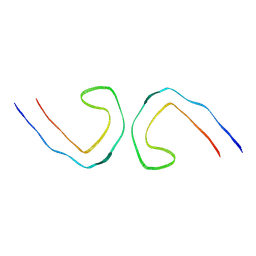 | | Cryo-EM structure of in vitro PHF fibril | | Descriptor: | Isoform Tau-D of Microtubule-associated protein tau | | Authors: | Li, X, Liu, C. | | Deposit date: | 2022-07-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Subtle change of fibrillation condition leads to substantial alteration of recombinant Tau fibril structure.
Iscience, 25, 2022
|
|
6N7X
 
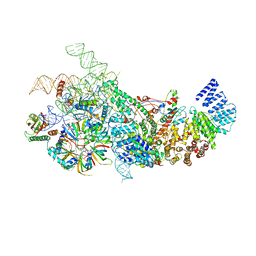 | | S. cerevisiae U1 snRNP | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, Pre-mRNA-processing factor 39, Protein NAM8, ... | | Authors: | Li, X, Liu, S, Jiang, J, Zhang, L, Espinosa, S, Hill, R.C, Hansen, K.C, Zhou, Z.H, Zhao, R. | | Deposit date: | 2018-11-28 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CryoEM structure of Saccharomyces cerevisiae U1 snRNP offers insight into alternative splicing.
Nat Commun, 8, 2017
|
|
7JI2
 
 | | Crystal Structure of H2-Kb in complex with a OVA mutant peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Li, X, Mallis, R.J, Mizsei, R, Tan, K, Reinherz, E.L, Wang, J. | | Deposit date: | 2020-07-22 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pre-T cell receptors topologically sample self-ligands during thymocyte beta-selection.
Science, 371, 2021
|
|
