3J8G
 
 | | Electron cryo-microscopy structure of EngA bound with the 50S ribosomal subunit | | Descriptor: | 23S rRNA, 50S ribosomal protein L1, 50S ribosomal protein L11, ... | | Authors: | Zhang, X, Yan, K, Zhang, Y, Li, N, Ma, C, Li, Z, Zhang, Y, Feng, B, Liu, J, Sun, Y, Xu, Y, Lei, J, Gao, N. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly
Nucleic Acids Res., 2014
|
|
1IAY
 
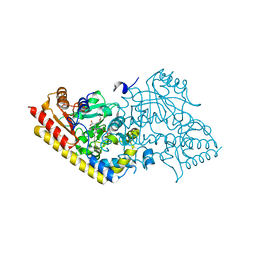 | | CRYSTAL STRUCTURE OF ACC SYNTHASE COMPLEXED WITH COFACTOR PLP AND INHIBITOR AVG | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE 2, 2-AMINO-4-(2-AMINO-ETHOXY)-BUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Huai, Q, Xia, Y, Chen, Y, Callahan, B, Li, N, Ke, H. | | Deposit date: | 2001-03-24 | | Release date: | 2001-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of 1-aminocyclopropane-1-carboxylate (ACC) synthase in complex with aminoethoxyvinylglycine and pyridoxal-5'-phosphate provide new insight into catalytic mechanisms
J.Biol.Chem., 276, 2001
|
|
1IAX
 
 | | CRYSTAL STRUCTURE OF ACC SYNTHASE COMPLEXED WITH PLP | | Descriptor: | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE 2, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Huai, Q, Xia, Y, Chen, Y, Callahan, B, Li, N, Ke, H. | | Deposit date: | 2001-03-24 | | Release date: | 2001-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of 1-aminocyclopropane-1-carboxylate (ACC) synthase in complex with aminoethoxyvinylglycine and pyridoxal-5'-phosphate provide new insight into catalytic mechanisms
J.Biol.Chem., 276, 2001
|
|
8FE5
 
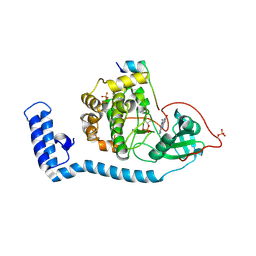 | | Structure of J-PKAc chimera complexed with Aplithianine B | | Descriptor: | 6-[(6P)-6-(1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-7,9-dihydro-8H-purin-8-one, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Du, L, Wilson, B.A.P, Li, N, Martinez Fiesco, J.A, Dalilian, M, Wang, D, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
5FTE
 
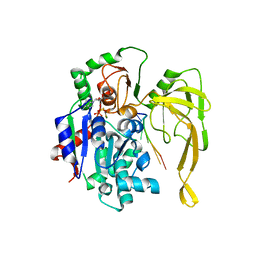 | | Crystal structure of Pif1 helicase from Bacteroides in complex with ADP-AlF3 and ssDNA | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP)-3', ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
1WZA
 
 | |
5FTC
 
 | | Crystal structure of Pif1 helicase from Bacteroides in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, TPR DOMAIN PROTEIN | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
5FTD
 
 | | Crystal structure of Pif1 helicase from Bacteroides apo form | | Descriptor: | PHOSPHATE ION, TPR DOMAIN PROTEIN | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
5FTF
 
 | | Crystal structure of Pif1 helicase from Bacteroides double mutant L95C-I339C | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TPR DOMAIN PROTEIN | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-13 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
5FTB
 
 | | Crystal structure of Pif1 helicase from Bacteroides in complex with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Chen, W.-F, Dai, Y.-X, Duan, X.-L, Liu, N.-N, Shi, W, Li, M, Dou, S.-X, Li, N, Dong, Y.-H, Rety, S, Xi, X.-G. | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal Structures of the Bspif1 Helicase Reveal that a Major Movement of the 2B SH3 Domain is Required for DNA Unwinding
Nucleic Acids Res., 44, 2016
|
|
3PFT
 
 | | Crystal Structure of Untagged C54A Mutant Flavin Reductase (DszD) in Complex with FMN From Mycobacterium goodii | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin reductase | | Authors: | Li, Q, Xu, P, Ma, C, Gu, L, Liu, X, Zhang, C, Li, N, Su, J, Li, B, Liu, S. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | The flavin reductase DSZD from a desulfurizing mycobacterium goodii strain: systemic manipulation and investigation based on the crystal structure
To be Published
|
|
5H5U
 
 | | Mechanistic insights into the alternative translation termination by ArfA and RF2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ma, C, Kurita, D, Li, N, Chen, Y, Himeno, H, Gao, N. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Mechanistic insights into the alternative translation termination by ArfA and RF2
Nature, 541, 2017
|
|
1AV2
 
 | | Gramicidin A/CsCl complex, active as a dimer | | Descriptor: | CESIUM ION, CHLORIDE ION, GRAMICIDIN A, ... | | Authors: | Burkhart, B.M, Li, N, Langs, D.A, Duax, W.L. | | Deposit date: | 1997-09-23 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Conducting Form of Gramicidin a is a Right-Handed Double-Stranded Double Helix.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BS9
 
 | | ACETYLXYLAN ESTERASE FROM P. PURPUROGENUM REFINED AT 1.10 ANGSTROMS | | Descriptor: | ACETYL XYLAN ESTERASE, SULFATE ION | | Authors: | Ghosh, D, Erman, M, Sawicki, M.W, Lala, P, Weeks, D.R, Li, N, Pangborn, W, Thiel, D.J, Jornvall, H, Eyzaguirre, J. | | Deposit date: | 1998-09-01 | | Release date: | 1999-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Determination of a protein structure by iodination: the structure of iodinated acetylxylan esterase.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
8WPF
 
 | | Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with exogenous DNA bearing one abasic site | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, A22R DNA polymerase processivity factor, DNA polymerase, ... | | Authors: | Wang, X, Li, N, Gao, N. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the assembly and mechanism of mpox virus DNA polymerase complex F8-A22-E4-H5.
Mol.Cell, 83, 2023
|
|
8WPE
 
 | | Structure of monkeypox virus polymerase complex F8-A22-E4-H5 (tag-free A22) with exogenous DNA | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, A22R DNA polymerase processivity factor, DNA polymerase, ... | | Authors: | Wang, X, Li, N, Gao, N. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the assembly and mechanism of mpox virus DNA polymerase complex F8-A22-E4-H5.
Mol.Cell, 83, 2023
|
|
8WPK
 
 | | Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with exgenous DNA | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA polymerase, DNA polymerase processivity factor, ... | | Authors: | Wang, X, Li, N, Gao, N. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the assembly and mechanism of mpox virus DNA polymerase complex F8-A22-E4-H5.
Mol.Cell, 83, 2023
|
|
8WPP
 
 | | Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with endogenous DNA | | Descriptor: | A22R DNA polymerase processivity factor, DNA polymerase, E4R Uracil-DNA glycosylase, ... | | Authors: | Wang, X, Li, N, Gao, N. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the assembly and mechanism of mpox virus DNA polymerase complex F8-A22-E4-H5.
Mol.Cell, 83, 2023
|
|
4GHQ
 
 | | Crystal structure of EV71 3C proteinase | | Descriptor: | 3C proteinase | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GHT
 
 | | Crystal structure of EV71 3C proteinase in complex with AG7088 | | Descriptor: | 3C proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8ERH
 
 | |
8ERE
 
 | |
8ERG
 
 | |
8ERI
 
 | |
8ERF
 
 | |
