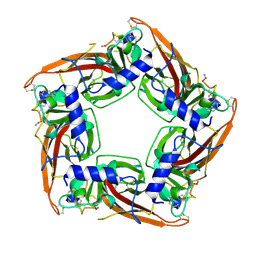
7N43
 
 | |
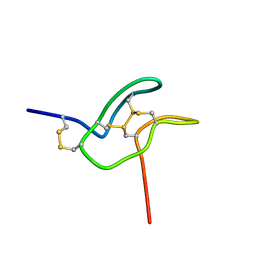
7SKC
 
 | |
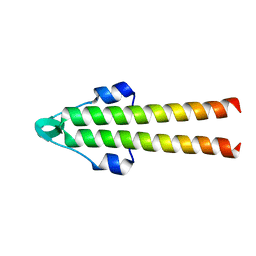
7O39
 
 | |
3QEF
 
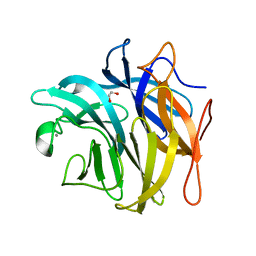 | | The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylosidase/alpha-L-arabinfuranosidase, gly43N, ... | | Authors: | Cartmell, A, Mckee, L.S, Pena, M, Larsbrink, J, Brumer, H, Lewis, R.J, Viks-Nielsen, A, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | The Structure and Function of an Arabinan-specific {alpha}-1,2-Arabinofuranosidase Identified from Screening the Activities of Bacterial GH43 Glycoside Hydrolases.
J.Biol.Chem., 286, 2011
|
|
7O61
 
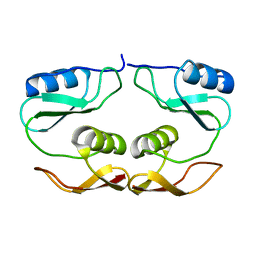 | |
3QEE
 
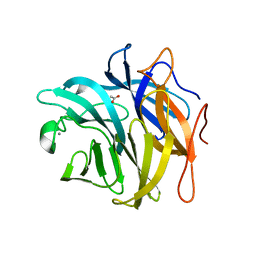 | | The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases | | Descriptor: | ACETATE ION, Beta-xylosidase/alpha-L-arabinfuranosidase, gly43N, ... | | Authors: | Cartmell, A, Mckee, L.S, Pena, M, Larsbrink, J, Brumer, H, Lewis, R.J, Viks-Nielsen, A, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Structure and Function of an Arabinan-specific {alpha}-1,2-Arabinofuranosidase Identified from Screening the Activities of Bacterial GH43 Glycoside Hydrolases.
J.Biol.Chem., 286, 2011
|
|
5AN5
 
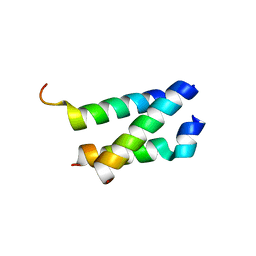 | | B. subtilis GpsB C-terminal Domain | | Descriptor: | CELL CYCLE PROTEIN GPSB, GLYCEROL | | Authors: | Rismondo, J, Cleverley, R.M, Lane, H.V, Grohennig, S, Steglich, A, Moller, L, Krishna Mannala, G, Hain, T, Lewis, R.J, Halbedel, S. | | Deposit date: | 2015-09-04 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the Bacterial Cell Division Determinant Gpsb and its Interaction with Penicillin Binding Proteins.
Mol.Microbiol., 99, 2016
|
|
2J6Y
 
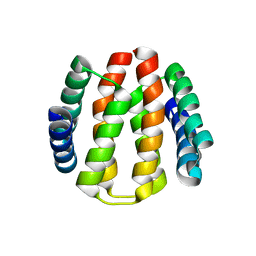 | | Structural and Functional Characterisation of partner switching regulating the environmental stress response in Bacillus subtilis | | Descriptor: | PHOSPHOSERINE PHOSPHATASE RSBU | | Authors: | Hardwick, S.W, Pane-Farre, J, Delumeau, O, Marles-Wright, J, Murray, J.W, Hecker, M, Lewis, R.J. | | Deposit date: | 2006-10-05 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional characterization of partner switching regulating the environmental stress response in Bacillus subtilis.
J. Biol. Chem., 282, 2007
|
|
2J6Z
 
 | | Structural and functional characterisation of partner-switching regulating the environmental stress response in B. subtilis | | Descriptor: | PHOSPHOSERINE PHOSPHATASE RSBU | | Authors: | Hardwick, S.W, Pane-Farre, J, Delumeau, O, Marles-Wright, J, Murray, J.W, Hecker, M, Lewis, R.J. | | Deposit date: | 2006-10-05 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and functional characterization of partner switching regulating the environmental stress response in Bacillus subtilis.
J. Biol. Chem., 282, 2007
|
|
2J70
 
 | | Structural and functional characterisation of partner-switching regulating the environmental stress response in B. subtilis | | Descriptor: | PHOSPHOSERINE PHOSPHATASE RSBU | | Authors: | Hardwick, S.W, Pane-Farre, J, Delumeau, O, Marles-Wright, J, Murray, J.W, Hecker, M, Lewis, R.J. | | Deposit date: | 2006-10-05 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and functional characterization of partner switching regulating the environmental stress response in Bacillus subtilis.
J. Biol. Chem., 282, 2007
|
|
2LR9
 
 | |
5FSR
 
 | | Crystal structure of penicillin binding protein 6B from Escherichia coli | | Descriptor: | D-ALANYL-D-ALANINE CARBOXYPEPTIDASE DACD | | Authors: | Peters, K, Kannan, S, Rao, V.A, Bilboy, J, Vollmer, D, Erickson, S.W, Lewis, R.J, Young, K.D, Vollmer, W. | | Deposit date: | 2016-01-07 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Redundancy of Peptidoglycan Carboxypeptidases Ensures Robust Cell Shape Maintenance in Escherichia Coli
Mbio, 7, 2016
|
|
4DE9
 
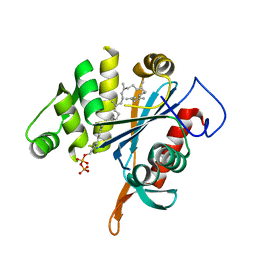 | | LytR-CPS2A-psr family protein YwtF (TagT) with bound octaprenyl pyrophosphate lipid | | Descriptor: | (2Z,6Z,10Z,14Z,18Z,22E,26E)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl trihydrogen diphosphate, Putative transcriptional regulator ywtF | | Authors: | Eberhardt, A, Hoyland, C.N, Vollmer, D, Bisle, S, Cleverley, R.M, Johnsborg, O, Havarstein, S, Lewis, R.J, Vollmer, W. | | Deposit date: | 2012-01-20 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | Attachment of Capsular Polysaccharide to the Cell Wall in Streptococcus pneumoniae.
Microb Drug Resist, 18, 2012
|
|
4HHF
 
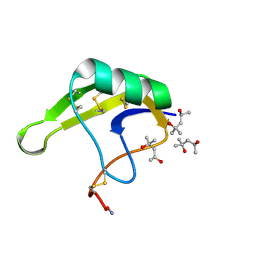 | | Crystal Structure of chemically synthesized scorpion alpha-toxin OD1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha-toxin OD1 | | Authors: | Wang, C.I.A, Lewis, R.J, Alewood, P.F, Durek, T. | | Deposit date: | 2012-10-09 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chemical engineering and structural and pharmacological characterization of the alpha-scorpion toxin OD1.
Acs Chem.Biol., 8, 2013
|
|
4I6O
 
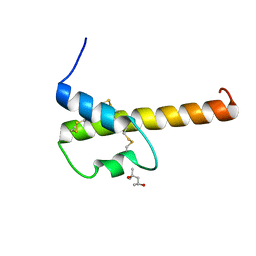 | | Crystal structure of chemically synthesized human anaphylatoxin C3a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Complement C3 | | Authors: | Wang, C.I.A, Ghassemian, A, Collins, B, Lewis, R.J, Alewood, P.F, Durek, T. | | Deposit date: | 2012-11-29 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Efficient chemical synthesis of human complement protein C3a.
Chem.Commun.(Camb.), 49, 2013
|
|
4OX5
 
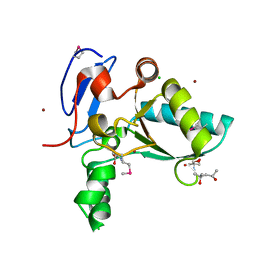 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-02-04 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
4OXD
 
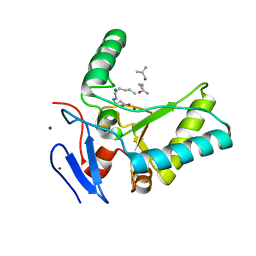 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | Descriptor: | CHLORIDE ION, LYSINE, LdcB LD-carboxypeptidase, ... | | Authors: | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-02-05 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
1W53
 
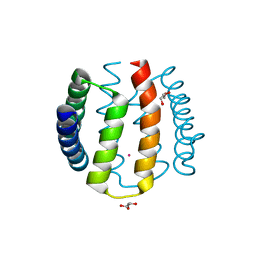 | | Kinase recruitment domain of the stress phosphatase RsbU | | Descriptor: | GLYCEROL, PHOSPHOSERINE PHOSPHATASE RSBU, XENON | | Authors: | Delumeau, O, Dutta, S, Brigulla, M, Kuhnke, G, Hardwick, S.W, Voelker, U, Yudkin, M.D, Lewis, R.J. | | Deposit date: | 2004-08-05 | | Release date: | 2004-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Functional and Structural Characterization of Rsbu, a Stress Signaling Protein Phosphatase 2C
J.Biol.Chem., 279, 2004
|
|
4OX3
 
 | | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition | | Descriptor: | PHOSPHATE ION, Putative carboxypeptidase YodJ, ZINC ION | | Authors: | Hoyland, C.N, Aldridge, C, Cleverley, R.M, Sidiq, K, Duchene, M.C, Daniel, R.A, Vollmer, W, Lewis, R.J. | | Deposit date: | 2014-02-04 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the LdcB LD-carboxypeptidase reveals the molecular basis of peptidoglycan recognition.
Structure, 22, 2014
|
|
1ONT
 
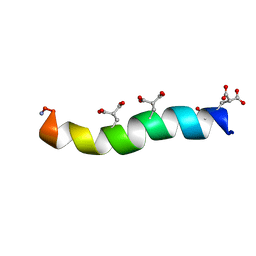 | | NMDA RECEPTOR ANTAGONIST, CONANTOKIN-T, NMR, 17 STRUCTURES | | Descriptor: | CONANTOKIN-T | | Authors: | Skjaerbaek, N, Nielsen, K.J, Lewis, R.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-08-27 | | Release date: | 1997-09-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of conantokin-G and conantokin-T by CD and NMR spectroscopy.
J.Biol.Chem., 272, 1997
|
|
1ONU
 
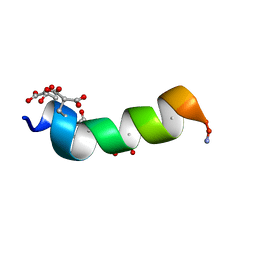 | | NMDA RECEPTOR ANTAGONIST, CONANTOKIN-G, NMR, 17 STRUCTURES | | Descriptor: | CONANTOKIN-G | | Authors: | Skjaerbaek, N, Nielsen, K.J, Lewis, R.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-08-27 | | Release date: | 1997-09-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of conantokin-G and conantokin-T by CD and NMR spectroscopy.
J.Biol.Chem., 272, 1997
|
|
1MTQ
 
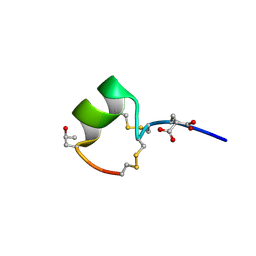 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF ALPHA-CONOTOXIN GID BY NMR SPECTROSCOPY | | Descriptor: | alpha-conotoxin GID | | Authors: | Nicke, A, Loughnan, M.L, Millard, E.L, Alewood, P.F, Adams, D.J, Daly, N.L, Craik, D.J, Lewis, R.J. | | Deposit date: | 2002-09-22 | | Release date: | 2003-02-11 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Isolation, Structure, and Activity of GID, a Novel alpha 4/7-Conotoxin with an Extended N-terminal Sequence
J.BIOL.CHEM., 278, 2003
|
|
1MVI
 
 | | N-TYPE CALCIUM CHANNEL BLOCKER, OMEGA-CONOTOXIN MVIIA, NMR, 15 STRUCTURES | | Descriptor: | MVIIA | | Authors: | Nielsen, K.J, Thomas, L, Lewis, R.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-08-02 | | Release date: | 1997-08-12 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | A consensus structure for omega-conotoxins with different selectivities for voltage-sensitive calcium channel subtypes: comparison of MVIIA, SVIB and SNX-202.
J.Mol.Biol., 263, 1996
|
|
1MVJ
 
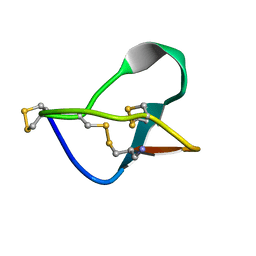 | | N-TYPE CALCIUM CHANNEL BLOCKER, OMEGA-CONOTOXIN MVIIA NMR, 15 STRUCTURES | | Descriptor: | SVIB | | Authors: | Nielsen, K.J, Thomas, L, Lewis, R.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-08-02 | | Release date: | 1997-08-12 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | A consensus structure for omega-conotoxins with different selectivities for voltage-sensitive calcium channel subtypes: comparison of MVIIA, SVIB and SNX-202.
J.Mol.Biol., 263, 1996
|
|
4AXO
 
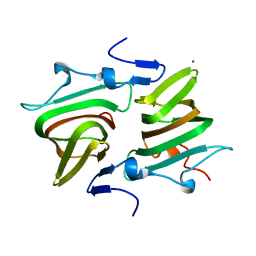 | | Structure of the Clostridium difficile EutQ protein | | Descriptor: | ETHANOLAMINE UTILIZATION PROTEIN, MAGNESIUM ION | | Authors: | Pitts, A.C, Tuck, L.R, Faulds-Pain, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2012-06-13 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Insight Into the Clostridium Difficile Ethanolamine Utilisation Microcompartment.
Plos One, 7, 2012
|
|
