2WI2
 
 | | Orally Active 2-Amino Thienopyrimidine Inhibitors of the Hsp90 Chaperone | | Descriptor: | 4-METHYL-6-(METHYLSULFANYL)-1,3,5-TRIAZIN-2-AMINE, DIMETHYL SULFOXIDE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Brough, P.A, Barril, X, Borgognoni, J, Chene, P, Davies, N.G.M, Davis, B, Drysdale, M.J, Dymock, B, Eccles, S.A, Garcia-Echeverria, C, Fromont, C, Hayes, A, Hubbard, R.E, Jordan, A.M, Rugaard-Jensen, M, Massey, A, Merret, A, Padfield, A, Parsons, R, Radimerski, T, Raynaud, F.I, Robertson, A, Roughley, S.D, Schoepfer, J, Simmonite, H, Surgenor, A, Valenti, M, Walls, S, Webb, P, Wood, M, Workman, P, Wright, L.M. | | Deposit date: | 2009-05-08 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Combining Hit Identification Strategies: Fragment- Based and in Silico Approaches to Orally Active 2-Aminothieno[2,3-D]Pyrimidine Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 52, 2009
|
|
2WI4
 
 | | Orally Active 2-Amino Thienopyrimidine Inhibitors of the Hsp90 Chaperone | | Descriptor: | 4-(2,4-dichlorophenyl)-5-phenyldiazenyl-pyrimidin-2-amine, HEAT SHOCK PROTEIN, HSP 90-ALPHA | | Authors: | Brough, P.A, Barril, X, Borgognoni, J, Chene, P, Davies, N.G.M, Davis, B, Drysdale, M.J, Dymock, B, Eccles, S.A, Garcia-Echeverria, C, Fromont, C, Hayes, A, Hubbard, R.E, Jordan, A.M, Rugaard-Jensen, M, Massey, A, Merret, A, Padfield, A, Parsons, R, Radimerski, T, Raynaud, F.I, Robertson, A, Roughley, S.D, Schoepfer, J, Simmonite, H, Surgenor, A, Valenti, M, Walls, S, Webb, P, Wood, M, Workman, P, Wright, L.M. | | Deposit date: | 2009-05-08 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Combining Hit Identification Strategies: Fragment- Based and in Silico Approaches to Orally Active 2-Aminothieno[2,3-D]Pyrimidine Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 52, 2009
|
|
6H5Q
 
 | | Cryo-EM structure of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to polyA RNA hexamers. | | Descriptor: | Nucleocapsid, RNA (5'-R(*AP*AP*AP*AP*AP*A)-3') | | Authors: | Desfosses, A, Milles, S, Ringkjobing Jensen, M, Guseva, S, Colletier, J, Maurin, D, Schoehn, G, Gutsche, I, Ruigrok, R, Blackledge, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6H5S
 
 | | Cryo-EM map of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to viral genomic 5-prime RNA hexamers. | | Descriptor: | Nucleocapsid, RNA (5'-R(*AP*CP*CP*AP*GP*A)-3') | | Authors: | Desfosses, A, Milles, S, Ringkjobing Jensen, M, Guseva, S, Colletier, J.P, Maurin, D, Schoehn, G, Gutsche, I, Ruigrok, R, Blackledge, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7PQK
 
 | |
7PKU
 
 | | Structure of SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a | | Descriptor: | 3C-like proteinase, Nucleoprotein | | Authors: | Bessa, L.M, Guseva, S, Camacho-Zarco, A.R, Salvi, N, Blackledge, M. | | Deposit date: | 2021-08-26 | | Release date: | 2022-01-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The intrinsically disordered SARS-CoV-2 nucleoprotein in dynamic complex with its viral partner nsp3a.
Sci Adv, 8, 2022
|
|
6FXA
 
 | |
3ZDO
 
 | | Tetramerization domain of Measles virus phosphoprotein | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PHOSPHOPROTEIN | | Authors: | Communie, G, Crepin, T, Jensen, M.R, Blackledge, M, Ruigrok, R.W.H. | | Deposit date: | 2012-11-29 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of the Tetramerization Domain of Measles Virus Phosphoprotein.
J.Virol., 87, 2013
|
|
4A56
 
 | | Crystal structure of the type 2 secretion system pilotin from Klebsiella Oxytoca | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PULLULANASE SECRETION PROTEIN PULS | | Authors: | Tosi, T, Nickerson, N.N, Mollica, L, RingkjobingJensen, M, Blackledge, M, Baron, B, England, P, Pugsley, A.P, Dessen, A. | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-07 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Pilotin-Secretin Recognition in the Type II Secretion System of Klebsiella Oxytoca.
Mol.Microbiol, 82, 2011
|
|
8POS
 
 | | Crystal structure of wolbachia leucyl-tRNA synthetase editing domain bound to cmpd9-AMP adduct | | Descriptor: | Leucine--tRNA ligase, SULFATE ION, [(1R,3'S,5S,6R,8R)-3'-(aminomethyl)-8-(6-aminopurin-9-yl)-4'-bromanyl-7'-[3-[methyl-(phenylmethyl)amino]propoxy]spiro[2,4,7-trioxa-3$l^{4}-borabicyclo[3.3.0]octane-3,1'-3H-2,1$l^{4}-benzoxaborole]-6-yl]methyl dihydrogen phosphate | | Authors: | Palencia, A, Hoffmann, G. | | Deposit date: | 2023-07-05 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | Targeting a microbiota Wolbachian aminoacyl-tRNA synthetase to block its pathogenic host.
Sci Adv, 10, 2024
|
|
8POT
 
 | | Ternary complex of E. coli leucyl-tRNA synthetase, tRNA(leu) and the benzoxaborole cmpd9 in the editing conformation | | Descriptor: | 1,2-ETHANEDIOL, Leucine--tRNA ligase, [(1R,3'S,5S,6R,8R)-3'-(aminomethyl)-8-(6-aminopurin-9-yl)-4'-bromanyl-7'-[3-[methyl-(phenylmethyl)amino]propoxy]spiro[2,4,7-trioxa-3$l^{4}-borabicyclo[3.3.0]octane-3,1'-3H-2,1$l^{4}-benzoxaborole]-6-yl]methyl dihydrogen phosphate, ... | | Authors: | Palencia, A, Hoffmann, G. | | Deposit date: | 2023-07-05 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Targeting a microbiota Wolbachian aminoacyl-tRNA synthetase to block its pathogenic host.
Sci Adv, 10, 2024
|
|
8POQ
 
 | |
8POR
 
 | |
3ZHM
 
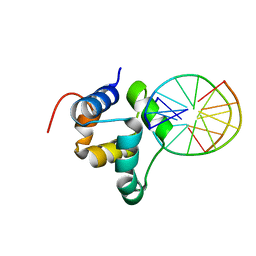 | | N-terminal domain of the CI repressor from bacteriophage TP901-1 in complex with the OL2 operator half-site | | Descriptor: | 5'-D(*AP*CP*GP*TP*GP*AP*AP*CP*TP*TP*GP*CP*AP*CP *TP*TP*GP*A)-3', 5'-D(*AP*GP*TP*TP*CP*AP*CP*GP*TP*TP*CP*AP*AP*GP *TP*GP*CP*A)-3', CI | | Authors: | Frandsen, K.H, Rasmussen, K.K, Poulsen, J.N, Lo Leggio, L. | | Deposit date: | 2012-12-22 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding of the N-Terminal Domain of the Lactococcal Bacteriophage Tp901-1 Ci Repressor to its Target DNA: A Crystallography, Small Angle Scattering, and Nuclear Magnetic Resonance Study.
Biochemistry, 52, 2013
|
|
3ZVU
 
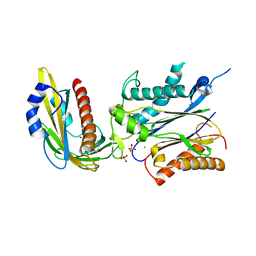 | | Structure of the PYR1 His60Pro mutant in complex with the HAB1 phosphatase and Abscisic acid | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABSCISIC ACID RECEPTOR PYR1, MANGANESE (II) ION, ... | | Authors: | Betz, K, Dupeux, F, Santiago, J, Rodriguez, P.L, Marquez, J.A. | | Deposit date: | 2011-07-27 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Thermodynamic Switch Modulates Abscisic Acid Receptor Sensitivity.
Embo J., 30, 2011
|
|
3ZBE
 
 | |
3ZHI
 
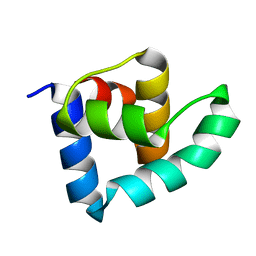 | | N-terminal domain of the CI repressor from bacteriophage TP901-1 | | Descriptor: | CI | | Authors: | Frandsen, K.H, Rasmussen, K.K, Poulsen, J.N, Lo Leggio, L. | | Deposit date: | 2012-12-21 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding of the N-Terminal Domain of the Lactococcal Bacteriophage Tp901-1 Ci Repressor to its Target DNA: A Crystallography, Small Angle Scattering, and Nuclear Magnetic Resonance Study.
Biochemistry, 52, 2013
|
|
4KTM
 
 | | Crystal Structure of C143S Xanthomonas campestris OleA | | Descriptor: | 3-oxoacyl-[ACP] synthase III, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION | | Authors: | Goblirsch, B.R. | | Deposit date: | 2013-05-20 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Substrate Trapping in Crystals of the Thiolase OleA Identifies Three Channels That Enable Long Chain Olefin Biosynthesis.
J.Biol.Chem., 291, 2016
|
|
4KTI
 
 | | Crystal Structure of C143A Xathomonas campestris OleA | | Descriptor: | 3-oxoacyl-[ACP] synthase III | | Authors: | Goblirsch, B.R. | | Deposit date: | 2013-05-20 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | Substrate Trapping in Crystals of the Thiolase OleA Identifies Three Channels That Enable Long Chain Olefin Biosynthesis.
J.Biol.Chem., 291, 2016
|
|
4KU5
 
 | |
4KU3
 
 | |
4KU2
 
 | |
5OC8
 
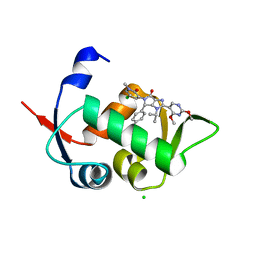 | | HDM2 (17-111, WILD TYPE) COMPLEXED WITH NVP-HDM201 AT 1.56A | | Descriptor: | (4~{S})-5-(5-chloranyl-1-methyl-2-oxidanylidene-pyridin-3-yl)-4-(4-chlorophenyl)-2-(2,4-dimethoxypyrimidin-5-yl)-3-propan-2-yl-4~{H}-pyrrolo[3,4-d]imidazol-6-one, CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Kallen, J. | | Deposit date: | 2017-06-29 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Dose and Schedule Determine Distinct Molecular Mechanisms Underlying the Efficacy of the p53-MDM2 Inhibitor HDM201.
Cancer Res., 78, 2018
|
|
3TT0
 
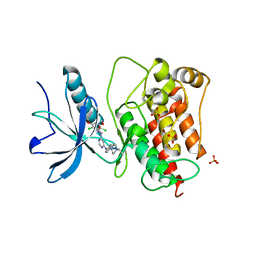 | | Co-structure of Fibroblast Growth Factor Receptor 1 kinase domain with 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (BGJ398) | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-(6-{[4-(4-ethylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)-1-methylurea, Basic fibroblast growth factor receptor 1, GLYCEROL, ... | | Authors: | Bussiere, D.E, Murray, J.M, Shu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase.
J.Med.Chem., 54, 2011
|
|
4HEO
 
 | |
