3SHO
 
 | |
3U7V
 
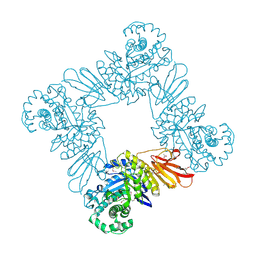 | |
3TEV
 
 | | The crystal structure of glycosyl hydrolase from Deinococcus radiodurans R1 | | Descriptor: | Glycosyl hyrolase, family 3 | | Authors: | Chang, C, Hatzos-Skintges, C, Kohler, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-08-15 | | Release date: | 2011-08-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of glycosyl hydrolase from Deinococcus radiodurans R1
To be Published
|
|
4HNH
 
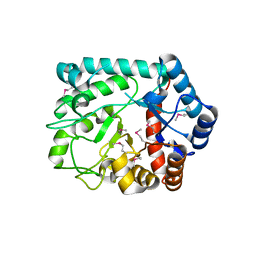
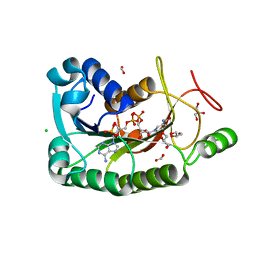 | | The crystal structure of a short-chain dehydrogenases/reductase (wide type) from Veillonella parvula DSM 2008 in complex with NADP | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-10-19 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.576 Å) | | Cite: | The crystal structure of a short-chain dehydrogenases/reductase (wide type) from Veillonella parvula DSM 2008 in complex with NADP.
To be Published
|
|
4HNG
 
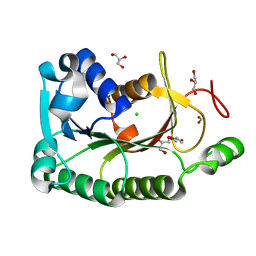 | | The crystal structure of a short-chain dehydrogenases/reductase (wide type) from Veillonella parvula DSM 2008 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-10-19 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a short-chain dehydrogenases/reductase (wide type) from Veillonella parvula DSM 2008
To be Published
|
|
4I4K
 
 | | Streptomyces globisporus C-1027 9-membered enediyne conserved protein SgcE6 | | Descriptor: | CITRIC ACID, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Kim, Y, Bigelow, L, Clancy, S, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J Antibiot (Tokyo), 69, 2016
|
|
4IAG
 
 | | Crystal structure of ZbmA, the zorbamycin binding protein from Streptomyces flavoviridis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Zbm binding protein | | Authors: | Cuff, M.E, Bigelow, L, Bruno, C.J.P, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-12-06 | | Release date: | 2013-02-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Zorbamycin-Binding Protein ZbmA, the Primary Self-Resistance Element in Streptomyces flavoviridis ATCC21892.
Biochemistry, 54, 2015
|
|
4HYL
 
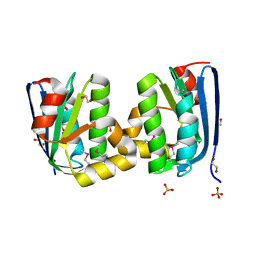 | | The crystal structure of an anti-sigma-factor antagonist from Haliangium ochraceum DSM 14365 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Stage II sporulation protein | | Authors: | Tan, K, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-13 | | Release date: | 2012-11-28 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The crystal structure of an anti-sigma-factor antagonist from Haliangium ochraceum DSM 14365
To be Published
|
|
2EW2
 
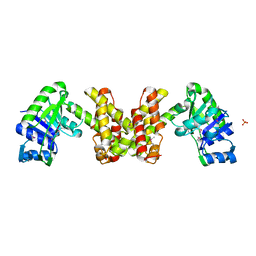 | | Crystal Structure of the Putative 2-Dehydropantoate 2-Reductase from Enterococcus faecalis | | Descriptor: | 2-dehydropantoate 2-reductase, putative, MAGNESIUM ION, ... | | Authors: | Kim, Y, Zhou, M, Moy, S, Clancy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-11-01 | | Release date: | 2005-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Putative 2-Dehydropantoate 2-Reductase from Enterococcus faecalis
To be Published
|
|
4ISC
 
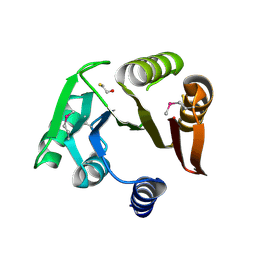 | | Crystal structure of a putative Methyltransferase from Pseudomonas syringae | | Descriptor: | BETA-MERCAPTOETHANOL, Methyltransferase | | Authors: | Filippova, E.V, Wawrzak, Z, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-16 | | Release date: | 2013-02-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of a putative Methyltransferase from Pseudomonas syringae
To be Published
|
|
4KQC
 
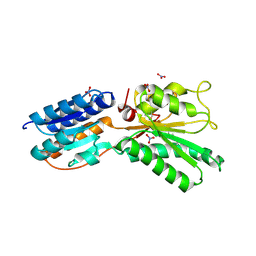 | | ABC transporter, LacI family transcriptional regulator from Brachyspira murdochii | | Descriptor: | NITRATE ION, Periplasmic binding protein/LacI transcriptional regulator | | Authors: | Osipiuk, J, Joachimiak, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-14 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | ABC transporter, LacI family transcriptional regulator from Brachyspira murdochii
To be Published
|
|
4IPT
 
 | | The crystal structure of a short-chain dehydrogenases/reductase (ethylated) from Veillonella parvula DSM 2008 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-10 | | Release date: | 2013-02-06 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.546 Å) | | Cite: | The crystal structure of a short-chain dehydrogenases/reductase (ethylated) from Veillonella parvula DSM 2008
To be Published
|
|
4KTB
 
 | | The crystal structure of posible asymmetric diadenosine tetraphosphate (Ap(4)A) hydrolases from Jonesia denitrificans DSM 20603 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative uncharacterized protein, ... | | Authors: | Tan, K, Kim, Y, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-20 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | The crystal structure of posible asymmetric diadenosine tetraphosphate (Ap(4)A) hydrolases from Jonesia denitrificans DSM 20603
To be Published
|
|
4KND
 
 | | Thioredoxin from Anaeromyxobacter dehalogenans. | | Descriptor: | Thioredoxin | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Clancy, S, Adkins, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2013-05-09 | | Release date: | 2013-05-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thioredoxin from Anaeromyxobacter dehalogenans.
To be Published
|
|
4I19
 
 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | Descriptor: | ACETATE ION, Epoxide hydrolase, FORMIC ACID | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
4EWF
 
 | |
4ESY
 
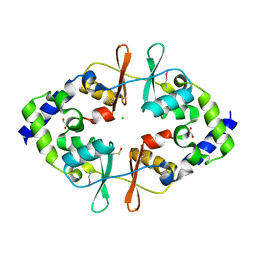 | | Crystal Structure of the CBS Domain of CBS Domain Containing Membrane Protein from Sphaerobacter thermophilus | | Descriptor: | 1,2-ETHANEDIOL, CBS domain containing membrane protein, CHLORIDE ION | | Authors: | Kim, Y, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-23 | | Release date: | 2012-09-05 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Crystal Structure of the CBS Domain of CBS Domain Containing Membrane Protein from Sphaerobacter thermophilus
To be Published
|
|
4DIM
 
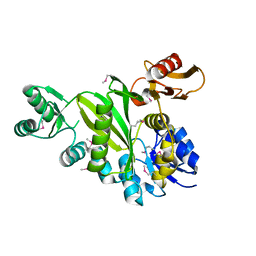 | |
4DQ0
 
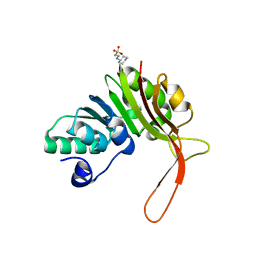 | |
4GBJ
 
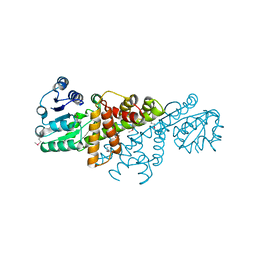 | |
4GS5
 
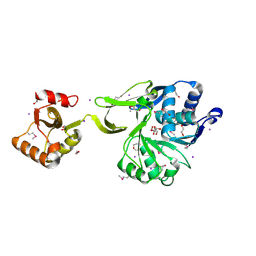 | | The crystal structure of acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II-like protein from Dyadobacter fermentans DSM 18053 | | Descriptor: | 1,2-ETHANEDIOL, Acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II-like protein, IODIDE ION | | Authors: | Tan, K, Holowicki, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-08-27 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.018 Å) | | Cite: | The crystal structure of acyl-CoA synthetase (AMP-forming)/AMP-acid ligase II-like protein from Dyadobacter fermentans DSM 18053
To be Published
|
|
4H3T
 
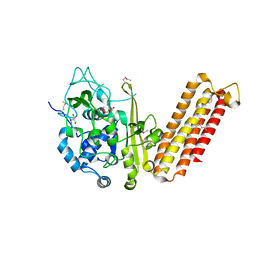 | | Crystal structure of CRISPR-associated protein Cse1 from Acidimicrobium ferrooxidans | | Descriptor: | CRISPR-associated protein, Cse1 family, GLYCEROL | | Authors: | Michalska, K, Stols, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-14 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of CRISPR-associated protein Cse1 from Acidimicrobium ferrooxidans
To be Published
|
|
4H7N
 
 | |
4HG2
 
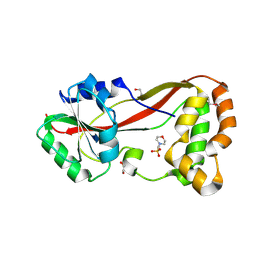 | | The Structure of a Putative Type II Methyltransferase from Anaeromyxobacter dehalogenans. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Methyltransferase type 11 | | Authors: | Cuff, M.E, Chhor, G, Clancy, S, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Adkins, J.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2012-10-05 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of a Putative Type II Methyltransferase from Anaeromyxobacter dehalogenans.
TO BE PUBLISHED
|
|
4H0C
 
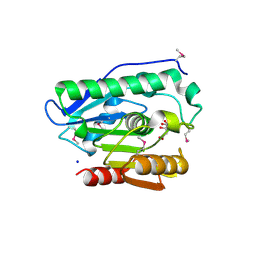 | | Crystal structure of phospholipase/Carboxylesterase from Dyadobacter fermentans DSM 18053 | | Descriptor: | CITRIC ACID, GLYCEROL, Phospholipase/Carboxylesterase, ... | | Authors: | Chang, C, Holowicki, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-07 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of phospholipase/Carboxylesterase from Dyadobacter fermentans DSM 18053
To be Published
|
|
