5Z8K
 
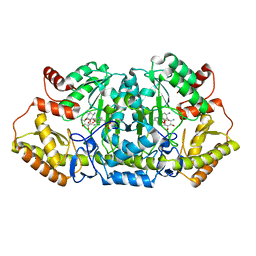 | | Crystal structure of an aminotransferase in complex with product-1 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, C-6' aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hong, S.K, Cha, S.S. | | Deposit date: | 2018-01-31 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Complete reconstitution of the diverse pathways of gentamicin B biosynthesis.
Nat. Chem. Biol., 15, 2019
|
|
5Z83
 
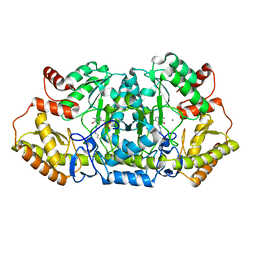 | |
5X9U
 
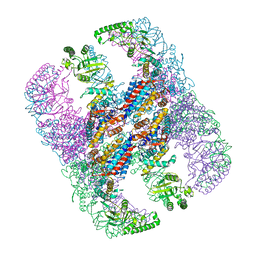 | | Crystal structure of group III chaperonin in the open state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thermosome, alpha subunit | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2017-03-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.001 Å) | | Cite: | Structural and mechanistic characterization of an archaeal-like chaperonin from a thermophilic bacterium
Nat Commun, 8, 2017
|
|
5H4U
 
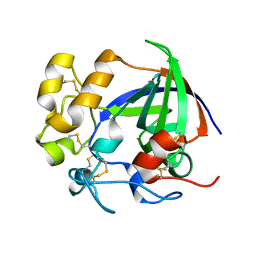 | | Crystal structure of cellulase from Antarctic springtail, Cryptopygus antarcticus | | Descriptor: | Endo-beta-1,4-glucanase | | Authors: | An, Y.J, Hong, S.K, Cha, S.S. | | Deposit date: | 2016-11-02 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Genetic and Structural Characterization of a Thermo-Tolerant, Cold-Active, and Acidic Endo-beta-1,4-glucanase from Antarctic Springtail, Cryptopygus antarcticus.
J. Agric. Food Chem., 65, 2017
|
|
5GSC
 
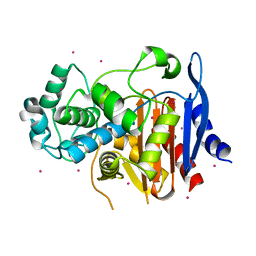 | |
5GZW
 
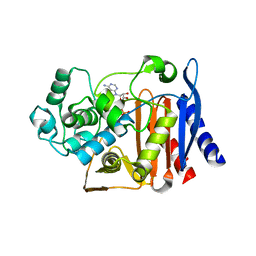 | | Crystal structure of AmpC BER adenylylated by acetyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase, SULFATE ION | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2016-10-02 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
5GGW
 
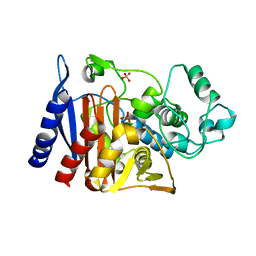 | | Crystal structure of Class C beta-lactamase | | Descriptor: | Beta-lactamase, PHOSPHATE ION, SULFATE ION | | Authors: | An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2016-06-16 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Structural basis for the extended substrate spectrum of AmpC BER and structure-guided discovery of the inhibition activity of citrate against the class C beta-lactamases AmpC BER and CMY-10.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2EWT
 
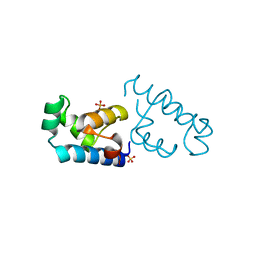 | | Crystal structure of the DNA-binding domain of BldD | | Descriptor: | SULFATE ION, putative DNA-binding protein | | Authors: | Kim, I.K, Lee, C.J, Kim, M.K, Kim, J.M, Kim, J.H, Yim, H.S, Cha, S.S, Kang, S.O. | | Deposit date: | 2005-11-07 | | Release date: | 2006-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of the DNA-binding domain of BldD, a central regulator of aerial mycelium formation in Streptomyces coelicolor A3(2)
Mol.Microbiol., 60, 2006
|
|
6KA5
 
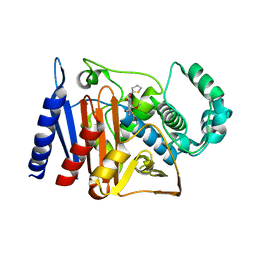 | | Crystal structure of a class C beta-lactamase in complex with cefoxitin | | Descriptor: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | Authors: | Bae, D.W, Jung, Y.E, An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2019-06-20 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Insights into Catalytic Relevances of Substrate Poses in ACC-1.
Antimicrob.Agents Chemother., 63, 2019
|
|
6K9T
 
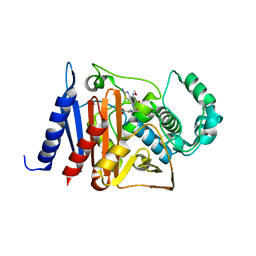 | | Crystal structure of a class C beta-lactamase in complex with cefotaxime | | Descriptor: | Beta-lactamase, CEFOTAXIME, C3' cleaved, ... | | Authors: | Bae, D.W, Jung, Y.E, An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2019-06-17 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.459 Å) | | Cite: | Structural Insights into Catalytic Relevances of Substrate Poses in ACC-1.
Antimicrob.Agents Chemother., 63, 2019
|
|
6KBY
 
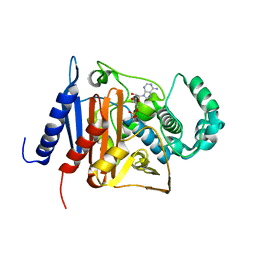 | | Crystal structure of a class C beta lactamase in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase | | Authors: | Bae, D.W, Jung, Y.E, An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-16 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | Structural Insights into Catalytic Relevances of Substrate Poses in ACC-1.
Antimicrob.Agents Chemother., 63, 2019
|
|
6K8X
 
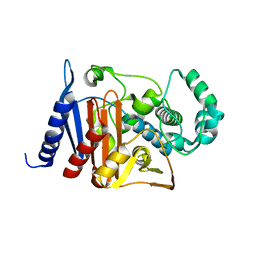 | | Crystal structure of a class C beta lactamase | | Descriptor: | Beta-lactamase | | Authors: | Bae, D.W, Jung, Y.E, An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2019-06-13 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Insights into Catalytic Relevances of Substrate Poses in ACC-1.
Antimicrob.Agents Chemother., 63, 2019
|
|
2BO1
 
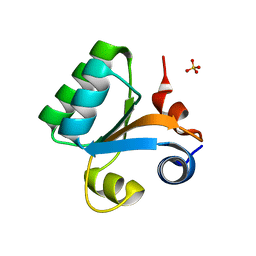 | |
1SMA
 
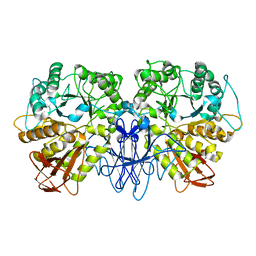 | | CRYSTAL STRUCTURE OF A MALTOGENIC AMYLASE | | Descriptor: | MALTOGENIC AMYLASE | | Authors: | Kim, J.S, Cha, S.S, Oh, B.H. | | Deposit date: | 1999-04-21 | | Release date: | 2000-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a maltogenic amylase provides insights into a catalytic versatility.
J.Biol.Chem., 274, 1999
|
|
2IP6
 
 | | Crystal structure of PedB | | Descriptor: | PapB, SULFATE ION | | Authors: | Kang, S.O, Kim, I.K, Kim, M.K, Kim, J.H, Yim, H.S, Cha, S.S. | | Deposit date: | 2006-10-12 | | Release date: | 2007-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High resolution crystal structure of PedB: a structural basis for the classification of pediocin-like immunity proteins
Bmc Struct.Biol., 7, 2007
|
|
3OUR
 
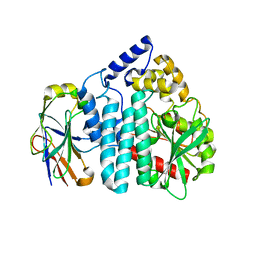 | |
3BP8
 
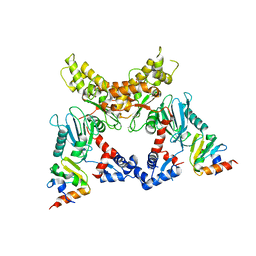 | | Crystal structure of Mlc/EIIB complex | | Descriptor: | ACETATE ION, PTS system glucose-specific EIICB component, Putative NAGC-like transcriptional regulator, ... | | Authors: | An, Y.J, Jung, H.I, Cha, S.S. | | Deposit date: | 2007-12-18 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Analyses of Mlc-IIBGlc interaction and a plausible molecular mechanism of Mlc inactivation by membrane sequestration
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZCU
 
 | | Crystal structure of a new type of NADPH-dependent quinone oxidoreductase (QOR2) from escherichia coli | | Descriptor: | COPPER (II) ION, Uncharacterized oxidoreductase ytfG | | Authors: | Kim, I.K, Yim, H.S, Kim, M.K, Kim, D.W, Kim, Y.M, Cha, S.S, Kang, S.O. | | Deposit date: | 2007-11-13 | | Release date: | 2008-05-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a new type of NADPH-dependent quinone oxidoreductase (QOR2) from Escherichia coli
J.Mol.Biol., 379, 2008
|
|
2ZCV
 
 | | Crystal structure of NADPH-dependent quinone oxidoreductase QOR2 complexed with NADPH from escherichia coli | | Descriptor: | COPPER (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Kim, I.K, Yim, H.S, Kim, M.K, Kim, D.W, Kim, Y.M, Cha, S.S, Kang, S.O. | | Deposit date: | 2007-11-13 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a new type of NADPH-dependent quinone oxidoreductase (QOR2) from Escherichia coli
J.Mol.Biol., 379, 2008
|
|
2HK0
 
 | | Crystal structure of D-psicose 3-epimerase (DPEase) in the absence of substrate | | Descriptor: | D-PSICOSE 3-EPIMERASE | | Authors: | Kim, K, Kim, H.J, Oh, D.K, Cha, S.S, Rhee, S. | | Deposit date: | 2006-07-03 | | Release date: | 2006-08-29 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of d-Psicose 3-epimerase from Agrobacterium tumefaciens and its Complex with True Substrate d-Fructose: A Pivotal Role of Metal in Catalysis, an Active Site for the Non-phosphorylated Substrate, and its Conformational Changes
J.Mol.Biol., 361, 2006
|
|
2HK1
 
 | | Crystal structure of D-psicose 3-epimerase (DPEase) in the presence of D-fructose | | Descriptor: | D-PSICOSE 3-EPIMERASE, D-fructose, MANGANESE (II) ION | | Authors: | Kim, K, Kim, H.J, Oh, D.K, Cha, S.S, Rhee, S. | | Deposit date: | 2006-07-03 | | Release date: | 2006-08-29 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of d-Psicose 3-epimerase from Agrobacterium tumefaciens and its Complex with True Substrate d-Fructose: A Pivotal Role of Metal in Catalysis, an Active Site for the Non-phosphorylated Substrate, and its Conformational Changes
J.Mol.Biol., 361, 2006
|
|
2JG7
 
 | | Crystal structure of Seabream Antiquitin and Elucidation of its substrate specificity | | Descriptor: | ANTIQUITIN, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tang, W.K, Wong, K.B, Cha, S.S, Lee, H.S, Cheng, C.H.K, Fong, W.P. | | Deposit date: | 2007-02-09 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | The Crystal Structure of Seabream Antiquitin Reveals the Structural Basis of its Substrate Specificity.
FEBS Lett., 582, 2008
|
|
4J4K
 
 | | Crystal structure of glucose isomerase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Xylose isomerase, ... | | Authors: | Kim, M.K, An, Y.J, Lee, S, Jeong, C.S, Cha, S.S. | | Deposit date: | 2013-02-07 | | Release date: | 2014-04-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of glucose isomerase
To be Published
|
|
2PQJ
 
 | | Crystal structure of active ribosome inactivating protein from maize (b-32), complex with adenine | | Descriptor: | ADENINE, Ribosome-inactivating protein 3 | | Authors: | Mak, A.N.S, Au, S.W.N, Cha, S.S, Young, J.A, Wong, K.B, Shaw, P.C. | | Deposit date: | 2007-05-02 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-function study of maize ribosome-inactivating protein: implications for the internal inactivation region and the sole glutamate in the active site.
Nucleic Acids Res., 35, 2007
|
|
2PQI
 
 | | Crystal structure of active ribosome inactivating protein from maize (b-32) | | Descriptor: | Ribosome-inactivating protein 3 | | Authors: | Mak, A.N.S, Wong, Y.T, Young, J.A, Cha, S.S, Sze, K.H, Au, S.W.N, Wong, K.B, Shaw, P.C. | | Deposit date: | 2007-05-02 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function study of maize ribosome-inactivating protein: implications for the internal inactivation region and the sole glutamate in the active site.
Nucleic Acids Res., 35, 2007
|
|
