7QEW
 
 | | human Connexin 26 class 2 hexamer at 90mmHg PCO2, pH7.4 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEQ
 
 | | human Connexin 26 dodecamer at 90mmHg PCO2, pH7.4 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEY
 
 | | human Connexin 26 class 1 hexamer at 90mmHg PCO2, pH7.4 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QER
 
 | | human Connexin 26 dodecamer at 55mm Hg PCO2, pH7.4 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEO
 
 | | human Connexin 26 at 55mm Hg PCO2, pH7.4: two masked subunits, class C | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEV
 
 | | human Connexin 26 at 55mm Hg PCO2, pH7.4:two masked subunits, class D | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QOA
 
 | | Structure of CodB, a cytosine transporter in an outward-facing conformation | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-AMINOPYRIMIDIN-2(1H)-ONE, Cytosine permease, ... | | Authors: | Hatton, C.E, Cameron, A.D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of cytosine transport protein CodB provides insight into nucleobase-cation symporter 1 mechanism.
Embo J., 41, 2022
|
|
4ATV
 
 | | STRUCTURE OF A TRIPLE MUTANT OF THE NHAA DIMER, CRYSTALLISED AT LOW PH | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, NA(+)/H(+) ANTIPORTER NHAA, SULFATE ION | | Authors: | Drew, D, Lee, C, Iwata, S, Cameron, A.D. | | Deposit date: | 2012-05-10 | | Release date: | 2013-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights.
J. Gen. Physiol., 144, 2014
|
|
7QEU
 
 | | human Connexin 26 at 55mmHg PCO2, pH7.4: two masked subunits, class B | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QES
 
 | | human Connexin 26 at 55mm Hg PCO2, pH7.4: two masked subunits, class A | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
4AU5
 
 | | Structure of the NhaA dimer, crystallised at low pH | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, NA(+)/H(+) ANTIPORTER NHAA, SULFATE ION | | Authors: | Drew, D, Lee, C, Iwata, S, Cameron, A.D. | | Deposit date: | 2012-05-14 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.696 Å) | | Cite: | Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights.
J. Gen. Physiol., 144, 2014
|
|
3ZUY
 
 | | Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Hu, N.-J, Iwata, S, Cameron, A.D, Drew, D. | | Deposit date: | 2011-07-21 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Bacterial Homologue of the Bile Acid Sodium Symporter Asbt.
Nature, 478, 2011
|
|
3ZUX
 
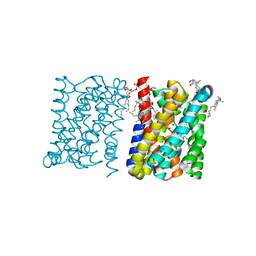 | | Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. | | Descriptor: | LAURYL DIMETHYLAMINE-N-OXIDE, MERCURY (II) ION, PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Hu, N.-J, Iwata, S, Cameron, A.D, Drew, D. | | Deposit date: | 2011-07-21 | | Release date: | 2011-10-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Bacterial Homologue of the Bile Acid Sodium Symporter Asbt.
Nature, 478, 2011
|
|
4BWZ
 
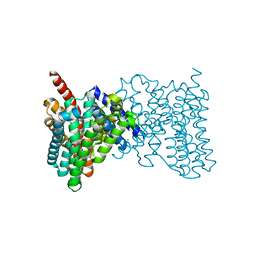 | |
1RK2
 
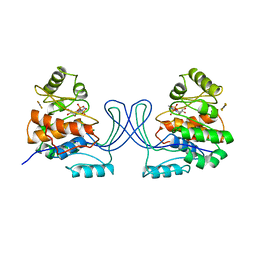 | | E. COLI RIBOKINASE COMPLEXED WITH RIBOSE AND ADP, SOLVED IN SPACE GROUP P212121 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RIBOKINASE, ... | | Authors: | Sigrell, J.A, Cameron, A.D, Mowbray, S.L. | | Deposit date: | 1999-05-20 | | Release date: | 1999-09-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Induced fit on sugar binding activates ribokinase.
J.Mol.Biol., 290, 1999
|
|
1RKS
 
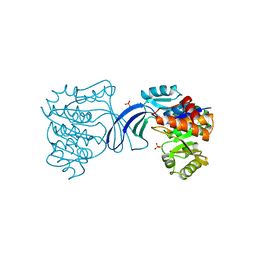 | |
1RKA
 
 | |
1RKD
 
 | | E. COLI RIBOKINASE COMPLEXED WITH RIBOSE AND ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, RIBOKINASE, ... | | Authors: | Sigrell, J.A, Cameron, A.D, Jones, T.A, Mowbray, S.L. | | Deposit date: | 1997-11-29 | | Release date: | 1998-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of Escherichia coli ribokinase in complex with ribose and dinucleotide determined to 1.8 A resolution: insights into a new family of kinase structures.
Structure, 6, 1998
|
|
2J4Z
 
 | | Structure of Aurora-2 in complex with PHA-680626 | | Descriptor: | 4-(4-METHYLPIPERAZIN-1-YL)-N-[5-(2-THIENYLACETYL)-1,5-DIHYDROPYRROLO[3,4-C]PYRAZOL-3-YL]BENZAMIDE, ARSENIC, SERINE THREONINE-PROTEIN KINASE 6 | | Authors: | Cameron, A.D, Izzo, G, Storici, P, Rusconi, L, Fancelli, D, Varasi, M, Berta, D, Bindi, S, Forte, B, Severino, D, Tonani, R, Vianello, P. | | Deposit date: | 2006-09-08 | | Release date: | 2006-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1,4,5,6-tetrahydropyrrolo[3,4-c]pyrazoles: identification of a potent Aurora kinase inhibitor with a favorable antitumor kinase inhibition profile.
J. Med. Chem., 49, 2006
|
|
2BMC
 
 | | Aurora-2 T287D T288D complexed with PHA-680632 | | Descriptor: | (3E)-N-(2,6-DIETHYLPHENYL)-3-{[4-(4-METHYLPIPERAZIN-1-YL)BENZOYL]IMINO}PYRROLO[3,4-C]PYRAZOLE-5(3H)-CARBOXAMIDE, SERINE THREONINE-PROTEIN KINASE 6 | | Authors: | Cameron, A.D, Izzo, G, Sagliano, A, Rusconi, L, Storici, P, Fancelli, D, Berta, D, Bindi, S, Catana, C, Forte, B, Giordano, P, Mantegani, S, Meroni, M, Moll, J, Pittala, V, Severino, D, Tonani, R, Varasi, M, Vulpetti, A, Vianello, P. | | Deposit date: | 2005-03-11 | | Release date: | 2005-03-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Potent and Selective Aurora Inhibitors Identified by the Expansion of a Novel Scaffold for Protein Kinase Inhibition.
J.Med.Chem., 48, 2005
|
|
6Y9C
 
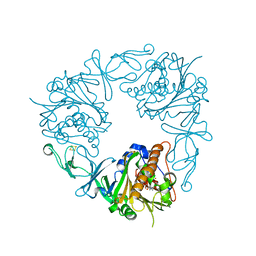 | | The structure of a quaternary ammonium Rieske monooxygenase reveals insights into carnitine oxidation by gut microbiota and inter-subunit electron transfer | | Descriptor: | CARNITINE, Carnitine monooxygenase oxygenase subunit, FE (III) ION, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterisation of an unusual cysteine pair in the Rieske carnitine monooxygenase CntA catalytic site.
Febs J., 2023
|
|
6ZGP
 
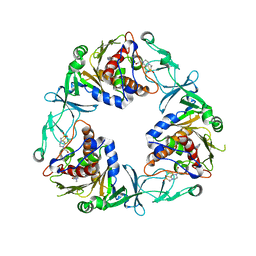 | | Crystal structure of the quaternary ammonium Rieske monooxygenase CntA in complex with inhibitor MMV12 (MMV020670) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Carnitine monooxygenase oxygenase subunit, FE (III) ION, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D.H, Cameron, A, Chen, Y. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
8QVN
 
 | |
6Y8J
 
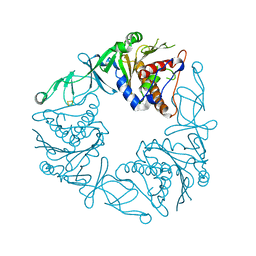 | | Crystal structure of the apo form of a quaternary ammonium Rieske monooxygenase CntA | | Descriptor: | Carnitine monooxygenase oxygenase subunit, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
6Y9D
 
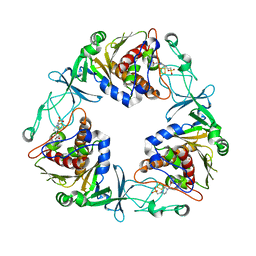 | | Crystal structure of the quaternary ammonium Rieske monooxygenase CntA in complex with substrate L-Carnitine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CARNITINE, Carnitine monooxygenase oxygenase subunit, ... | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-06 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
