6U8F
 
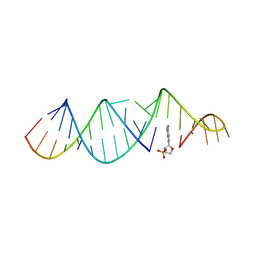 | | RNA hairpin, bound with TNA monomer | | Descriptor: | 2-azanyl-9-[(2~{R},3~{R},4~{S})-3-oxidanyl-4-[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxy-oxolan-2-yl]-1~{H}-purin-6-one, RNA (5'-R(*CP*UP*GP*CP*UP*GP*GP*CP*UP*AP*AP*GP*GP*CP*AP*UP*GP*AP*AP*AP*GP*U)-3'), RNA (5'-R(P*CP*UP*AP*UP*GP*CP*CP*UP*GP*CP*UP*G)-3') | | Authors: | Szostak, J.W, Zhang, W. | | Deposit date: | 2019-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural interpretation of the effects of threo-nucleotides on nonenzymatic template-directed polymerization.
Nucleic Acids Res., 49, 2021
|
|
6U7Y
 
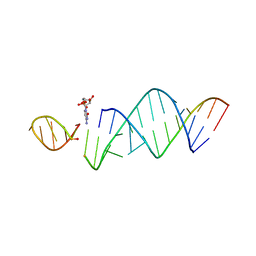 | | RNA hairpin structure containing one TNA nucleotide as template | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, RNA (5'-R(*CP*UP*AP*UP*GP*CP*CP*UP*GP*CP*UP*G)-3'), RNA (5'-R(*CP*UP*GP*CP*UP*GP*GP*CP*UP*AP*AP*GP*GP*(TG)P*CP*CP*GP*AP*AP*AP*GP*G)-3') | | Authors: | Szostak, J.W, Zhang, W. | | Deposit date: | 2019-09-03 | | Release date: | 2020-12-09 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural interpretation of the effects of threo-nucleotides on nonenzymatic template-directed polymerization.
Nucleic Acids Res., 49, 2021
|
|
7XKY
 
 | |
5DNI
 
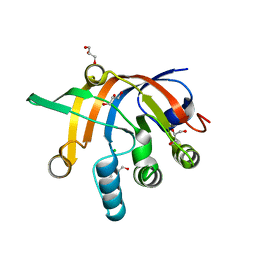 | | Crystal structure of Methanocaldococcus jannaschii Fumarate hydratase beta subunit | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jayaraman, V, Kunala, J, Balaram, H. | | Deposit date: | 2015-09-10 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Revisiting the Burden Borne by Fumarase: Enzymatic Hydration of an Olefin.
Biochemistry, 62, 2023
|
|
7D95
 
 | |
7D40
 
 | |
7D97
 
 | |
7D96
 
 | |
3JQJ
 
 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 | | Descriptor: | GLYCEROL, Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION, ... | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Nakagawa, N, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3JQK
 
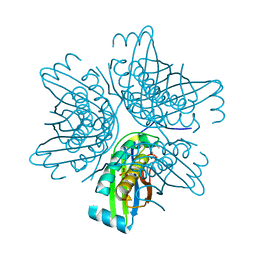 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 (H32 FORM) | | Descriptor: | ACETATE ION, Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Nakagawa, N, Sekar, K, Baba, S, Chen, L, Liu, Z.-J, Wang, B.-C, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6P78
 
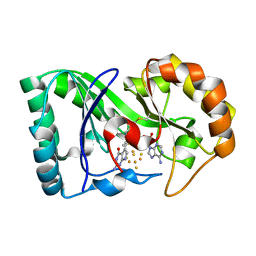 | | queuine lyase from Clostridium spiroforme bound to SAM and queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5S)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-1,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, IRON/SULFUR CLUSTER, Queuine lyase, ... | | Authors: | Almo, S.C, Grove, T.L. | | Deposit date: | 2019-06-05 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.726 Å) | | Cite: | Discovery of novel bacterial queuine salvage enzymes and pathways in human pathogens.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8T4S
 
 | |
3JQM
 
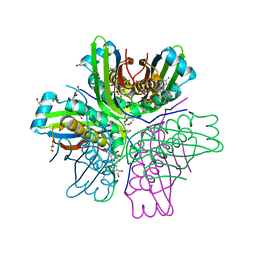 | | Binding of 5'-GTP to molybdenum cofactor biosynthesis protein MoaC from Thermus theromophilus HB8 | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Nakagawa, N, Sekar, K, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 66, 2010
|
|
7F32
 
 | |
5VHE
 
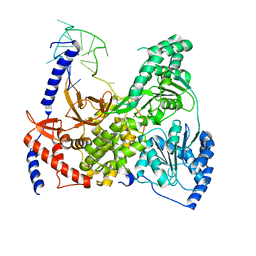 | | DHX36 in complex with the c-Myc G-quadruplex | | Descriptor: | DEAH (Asp-Glu-Ala-His) box polypeptide 36, DNA (5'-D(*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*TP*TP*TP*TP*TP*T)-3'), POTASSIUM ION | | Authors: | Chen, M, Ferre-D'Amare, A. | | Deposit date: | 2017-04-13 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.793 Å) | | Cite: | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
5VHA
 
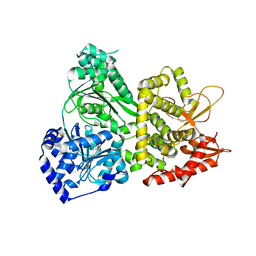 | | DHX36 with an N-terminal truncation | | Descriptor: | DEAH (Asp-Glu-Ala-His) box polypeptide 36 | | Authors: | Chen, M, Ferre-D'Amare, A. | | Deposit date: | 2017-04-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
5VHD
 
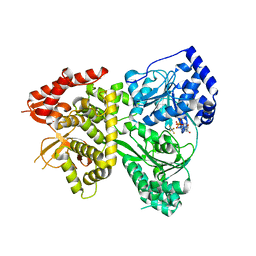 | | DHX36 with an N-terminal truncation bound to ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DEAH (Asp-Glu-Ala-His) box polypeptide 36, TETRAFLUOROALUMINATE ION | | Authors: | Chen, M, Ferre-D'Amare, A. | | Deposit date: | 2017-04-13 | | Release date: | 2018-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
5VHC
 
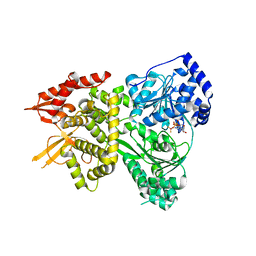 | | DHX36 with an N-terminal truncation bound to ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DEAH (Asp-Glu-Ala-His) box polypeptide 36, ... | | Authors: | Chen, M, Ferre-D'Amare, A. | | Deposit date: | 2017-04-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36.
Nature, 558, 2018
|
|
5LS6
 
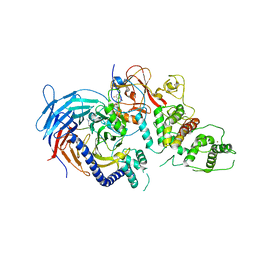 | | Structure of Human Polycomb Repressive Complex 2 (PRC2) with inhibitor | | Descriptor: | 1-[(1~{R})-1-[1-[2,2-bis(fluoranyl)propyl]piperidin-4-yl]ethyl]-~{N}-[(4-methoxy-6-methyl-2-oxidanylidene-3~{H}-pyridin-3-yl)methyl]-2-methyl-indole-3-carboxamide, Histone-lysine N-methyltransferase EZH2,Histone-lysine N-methyltransferase EZH2,Histone-lysine N-methyltransferase EZH2, Jarid2 K116me3, ... | | Authors: | Zhang, Y, Justin, N, Chen, S, Wilson, J, Gamblin, S. | | Deposit date: | 2016-08-22 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Identification of (R)-N-((4-Methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a Potent and Selective Inhibitor of Histone Methyltransferase EZH2, Suitable for Phase I Clinical Trials for B-Cell Lymphomas.
J. Med. Chem., 59, 2016
|
|
6JVB
 
 | |
6JV9
 
 | | Crystal Structure of Human CRMP2 1-532, unmodified | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Dihydropyrimidinase-related protein 2, ... | | Authors: | Jiang, X, Ogawa, T, Hirokawa, N. | | Deposit date: | 2019-04-16 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Enhanced carbonyl stress induces irreversible multimerization of CRMP2 in schizophrenia pathogenesis.
Life Sci Alliance, 2, 2019
|
|
3ZEV
 
 | | Structure of Thermostable Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | GLYCINE, NEUROTENSIN, NEUROTENSIN RECEPTOR 1 TM86V | | Authors: | Egloff, P, Hillenbrand, M, Schlinkmann, K.M, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2012-12-07 | | Release date: | 2014-01-29 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7U0H
 
 | | State NE1 nucleolar 60S ribosome biogenesis intermediate - Overall model | | Descriptor: | 25S rRNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, 5.8S rRNA, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2022-02-18 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4BUO
 
 | | High Resolution Structure of Thermostable Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | GLYCINE, NEUROTENSIN RECEPTOR TYPE 1, NEUROTENSIN/NEUROMEDIN N | | Authors: | Egloff, P, Hillenbrand, M, Schlinkmann, K.M, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2013-06-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7PNB
 
 | | Sulfolobus acidocaldarius 0406 filament. | | Descriptor: | 6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-[alpha-D-mannopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sulfolobus acidocaldarius 0406 filament., beta-D-glucopyranose-(1-4)-6-deoxy-6-sulfo-beta-D-glucopyranose-(1-3)-[alpha-D-mannopyranose-(1-4)][alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Isupov, M.N, Gaines, M, Daum, B. | | Deposit date: | 2021-09-06 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Electron cryo-microscopy reveals the structure of the archaeal thread filament.
Nat Commun, 13, 2022
|
|
