6UX6
 
 | | Cruzain covalently bound by a vinylsulfone compound | | Descriptor: | Cruzipain, GLYCEROL, Nalpha-[(benzyloxy)carbonyl]-N-[(2S)-1-phenyl-4-(phenylsulfonyl)butan-2-yl]-L-phenylalaninamide | | Authors: | Zhai, X, Tang, S, Chenna, B.C, Meek, T.D, Sacchettini, J.C. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Cruzain covalently bound by a vinylsulfone compound
To Be Published
|
|
6VB9
 
 | | Covalent adduct of cis-2,3-epoxysuccinic acid with Isocitrate Lyase-1 from Mycobacterium tuberculosis | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, Isocitrate lyase, ... | | Authors: | Krieger, I.V, Pham, T.V, Meek, T.D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Covalent Inactivation of Mycobacterium tuberculosis Isocitrate Lyase by cis -2,3-Epoxy-Succinic Acid.
Acs Chem.Biol., 16, 2021
|
|
4WIE
 
 | |
8SUT
 
 | | Crystal structure of YisK from Bacillus subtilis in complex with reaction product 4-Hydroxy-2-oxoglutaric acid | | Descriptor: | (2~{R})-2-oxidanyl-4-oxidanylidene-pentanedioic acid, Fumarylacetoacetate hydrolase family protein, MANGANESE (II) ION | | Authors: | Krieger, I.V, Chemelewski, V, Guo, T, Sperber, A, Herman, J, Sacchettini, J.C. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Bacillus subtilis YisK possesses oxaloacetate decarboxylase activity and exhibits Mbl-dependent localization.
J.Bacteriol., 206, 2024
|
|
8SKY
 
 | | Crystal structure of YisK from Bacillus subtilis in complex with oxalate | | Descriptor: | Fumarylacetoacetate hydrolase family protein, MANGANESE (II) ION, OXALATE ION | | Authors: | Krieger, I.V, Chemelewski, V, Guo, T, Sperber, A, Herman, J, Sacchettini, J.C. | | Deposit date: | 2023-04-20 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacillus subtilis YisK possesses oxaloacetate decarboxylase activity and exhibits Mbl-dependent localization.
J.Bacteriol., 206, 2024
|
|
8SUU
 
 | | Crystal structure of YisK from Bacillus subtilis in apo form | | Descriptor: | Fumarylacetoacetate hydrolase family protein, MANGANESE (II) ION | | Authors: | Krieger, I.V, Chemelewski, V, Guo, T, Sperber, A, Herman, J, Sacchettini, J.C. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Bacillus subtilis YisK possesses oxaloacetate decarboxylase activity and exhibits Mbl-dependent localization.
J.Bacteriol., 206, 2024
|
|
4WPV
 
 | |
4WPT
 
 | |
4WPU
 
 | |
4WOU
 
 | |
5KVV
 
 | |
7RB1
 
 | | Isocitrate Lyase-1 from Mycobacterium tuberculosis covalently modified by 5-descarboxy-5-nitro-D-isocitric acid | | Descriptor: | (3E)-3-(hydroxyimino)propanoic acid, GLYCEROL, GLYOXYLIC ACID, ... | | Authors: | Krieger, I.V, Mellott, D, Meek, T, Sacchettini, J.C. | | Deposit date: | 2021-07-05 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism-Based Inactivation of Mycobacterium tuberculosis Isocitrate Lyase 1 by (2 R ,3 S )-2-Hydroxy-3-(nitromethyl)succinic acid.
J.Am.Chem.Soc., 143, 2021
|
|
7S8W
 
 | | Amycolatopsis sp. T-1-60 N-succinylamino acid racemase/o-succinylbenzoate synthase R266Q mutant in complex with N-succinylphenylglycine | | Descriptor: | MAGNESIUM ION, N-succinyl-L-phenylglycine, N-succinylamino acid racemase/O-succinylbenzoate synthase, ... | | Authors: | Truong, D.P, Rousseau, S, Sacchettini, J.C, Glasner, M.E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Second-Shell Amino Acid R266 Helps Determine N -Succinylamino Acid Racemase Reaction Specificity in Promiscuous N -Succinylamino Acid Racemase/ o -Succinylbenzoate Synthase Enzymes.
Biochemistry, 60, 2021
|
|
8TQV
 
 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with inhibitor X20403 | | Descriptor: | 4-(2-{(4M)-4-[(6M)-6-(2,5-dimethoxyphenyl)pyridin-3-yl]-1H-1,2,3-triazol-1-yl}ethyl)-N-{[1-(methoxymethyl)cyclopropyl]methyl}-N-methylbenzamide, Polyketide synthase Pks13, SULFATE ION | | Authors: | Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2023-08-08 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitors of the Thioesterase Activity of Mycobacterium tuberculosis Pks13 Discovered Using DNA-Encoded Chemical Library Screening.
Acs Infect Dis., 10, 2024
|
|
7RUR
 
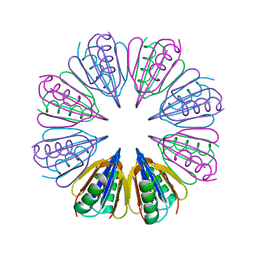 | |
7RUS
 
 | |
8TRY
 
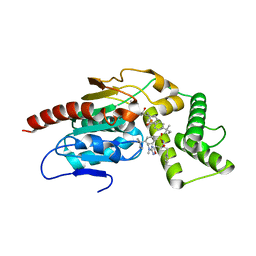 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with inhibitor X20348 | | Descriptor: | N-{(2S,3S)-4-[3-(dimethylamino)-1,2,4-oxadiazol-5-yl]-3-hydroxy-1-phenylbutan-2-yl}-4-(2-methylbutan-2-yl)benzene-1-sulfonamide, Polyketide synthase Pks13, SULFATE ION | | Authors: | Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2023-08-10 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Inhibitors of the Thioesterase Activity of Mycobacterium tuberculosis Pks13 Discovered Using DNA-Encoded Chemical Library Screening.
Acs Infect Dis., 10, 2024
|
|
8TR4
 
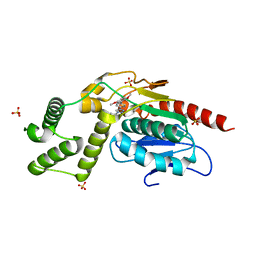 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with inhibitor X20404 | | Descriptor: | 4-(2-{(4M)-4-[(6M)-6-(2,5-dimethoxyphenyl)pyridin-3-yl]-1H-1,2,3-triazol-1-yl}ethyl)-N,N-dimethylbenzamide, Polyketide synthase Pks13, SULFATE ION | | Authors: | Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of the Thioesterase Activity of Mycobacterium tuberculosis Pks13 Discovered Using DNA-Encoded Chemical Library Screening.
Acs Infect Dis., 10, 2024
|
|
6PK2
 
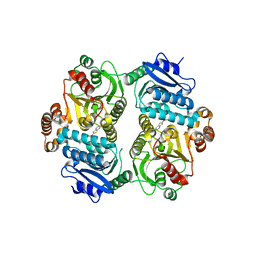 | | CRYSTAL STRUCTURE OF THE CARBOXYLTRANSFERASE SUBUNIT OF ACC (ACCD6) IN COMPLEX WITH INHIBITOR QUIZALOFOP-P derivative FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | 2-{4-[(6-fluoro-1,3-benzothiazol-2-yl)oxy]-2-hydroxyphenyl}-N-methylacetamide, Propionyl-CoA carboxylase subunit beta | | Authors: | Reddy, M.C.M, Nian, Z, Michele, T.C.B, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Elucidating the Inhibition and specificity of binding of
herbicidal aryloxyphenoxypropionates derivatives to Mycobacterium tuberculosis carboxyltransferase
domain of acetyl-coenzyme A(AccD6).
To Be Published
|
|
6P7U
 
 | |
6PRW
 
 | |
1KZX
 
 | | Solution structure of human intestinal fatty acid binding protein with a naturally-occurring single amino acid substitution (A54T) | | Descriptor: | INTESTINAL FATTY ACID-BINDING PROTEIN (T54) | | Authors: | Zhang, F, Luecke, C, Baier, L.J, Sacchettini, J.C, Hamilton, J.A. | | Deposit date: | 2002-02-08 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human intestinal fatty acid binding protein with a naturally-occurring single amino acid substitution (A54T) that is associated with altered lipid metabolism
Biochemistry, 42, 2003
|
|
1KZW
 
 | | Solution structure of Human Intestinal Fatty acid binding protein | | Descriptor: | INTESTINAL FATTY ACID-BINDING PROTEIN (A54) | | Authors: | Zhang, F, Luecke, C, Baier, L.J, Sacchettini, J.C, Hamilton, J.A. | | Deposit date: | 2002-02-08 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human intestinal fatty acid binding protein with a naturally-occurring single amino acid substitution (A54T) that is associated with altered lipid metabolism
Biochemistry, 42, 2003
|
|
2ZJ1
 
 | | Crystal structure of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with NAD and 3'-keto-aristeromycin | | Descriptor: | (2S,3R,5R)-3-(6-amino-9H-purin-9-yl)-2-hydroxy-5-(hydroxymethyl)cyclopentanone, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Reddy, M.C.M, Gokulan, K, Shetty, N.D, Owen, J.L, Ioerger, T.R, Sacchettini, J.C. | | Deposit date: | 2008-02-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with substrate and inhibitors.
Protein Sci., 17, 2008
|
|
2ZJ0
 
 | | Crystal structure of Mycobacterium tuberculosis S-Adenosyl-L-homocysteine hydrolase in ternary complex with NAD and 2-fluoroadenosine | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Reddy, M.C.M, Gokulan, K, Shetty, N.D, Owen, J.L, Ioerger, T.R, Sacchettini, J.C. | | Deposit date: | 2008-02-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with substrate and inhibitors.
Protein Sci., 17, 2008
|
|
