8CYJ
 
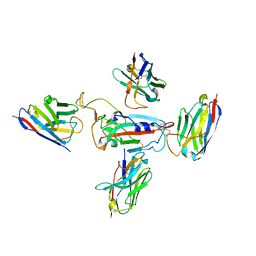 | | RBD of SARS-CoV-2 Spike protein in complex with pan-sarbecovirus nanobodies 2-10, 2-67, 2-62 and 1-25 | | Descriptor: | Spike glycoprotein, pan-sarbecovirus nanobody 1-25, pan-sarbecovirus nanobody 2-10, ... | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2022-05-23 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Superimmunity by pan-sarbecovirus nanobodies.
Cell Rep, 39, 2022
|
|
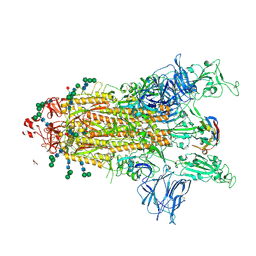
8CYB
 
 | |
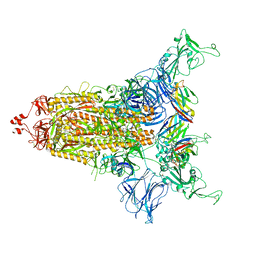
8CYD
 
 | |
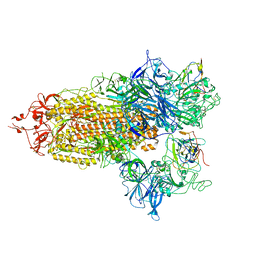
8CXN
 
 | |
8CY9
 
 | |
8CY7
 
 | |
8E3S
 
 | |
6UP7
 
 | | neurotensin receptor and arrestin2 complex | | Descriptor: | ARG-ARG-PRO-TYR-ILE-LEU, Beta-arrestin-1, Neurotensin receptor type 1, ... | | Authors: | Qu, Q.H, Huang, W, Masureel, M, Janetzko, J, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2019-10-16 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the neurotensin receptor 1 in complex with beta-arrestin 1.
Nature, 579, 2020
|
|
7SBF
 
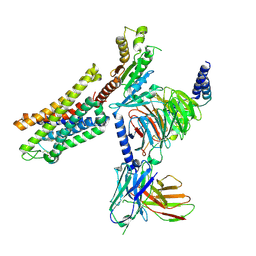 | | PZM21 bound Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Huang, W, Qu, Q, Wang, H, Skiniotis, G, Kobilka, B. | | Deposit date: | 2021-09-24 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-Based Evolution of G Protein-Biased mu-Opioid Receptor Agonists.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7N9T
 
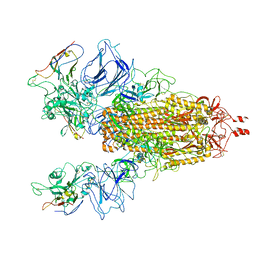 | | CryoEM structure of SARS-CoV-2 Spike in complex with Nb17 | | Descriptor: | Nanobody Nb17, Spike glycoprotein | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2021-06-18 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Potent neutralizing nanobodies resist convergent circulating variants of SARS-CoV-2 by targeting diverse and conserved epitopes
Nat Commun, 12, 2021
|
|
7MEJ
 
 | |
7MDW
 
 | |
7ME7
 
 | |
6NTS
 
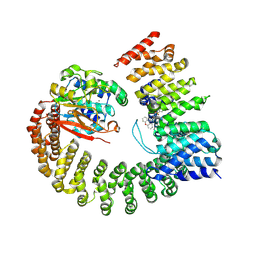 | | Protein Phosphatase 2A (Aalpha-B56alpha-Calpha) holoenzyme in complex with a Small Molecule Activator of PP2A (SMAP) | | Descriptor: | MANGANESE (II) ION, N-[(1R,2R,3S)-2-hydroxy-3-(10H-phenoxazin-10-yl)cyclohexyl]-4-(trifluoromethoxy)benzene-1-sulfonamide, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform, ... | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2019-01-30 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Selective PP2A Enhancement through Biased Heterotrimer Stabilization.
Cell, 181, 2020
|
|
8FZR
 
 | |
8INA
 
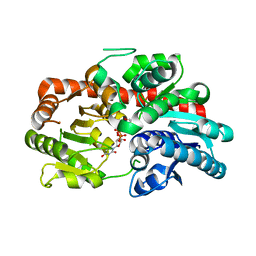 | | Crystal structure of UGT74AN3-UDP | | Descriptor: | GLYCEROL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Long, F, Huang, W. | | Deposit date: | 2023-03-09 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8INV
 
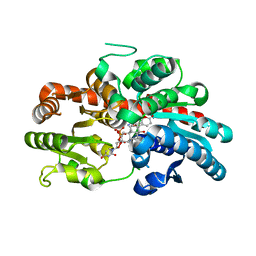 | | Crystal structure of UGT74AN3-UDP-BUF | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Long, F, Huang, W. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8INO
 
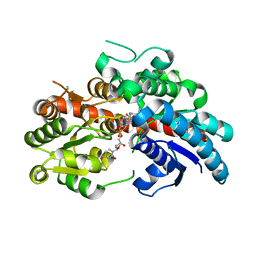 | | Crystal structure of UGT74AN3 in complex UDP and PER | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(3S,5S,8S,9S,10R,13R,14S,17R)-10,13-dimethyl-3,5,14-tris(oxidanyl)-2,3,4,6,7,8,9,11,12,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2H-furan-5-one, Glycosyltransferase, ... | | Authors: | Long, F, Huang, W. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8K5U
 
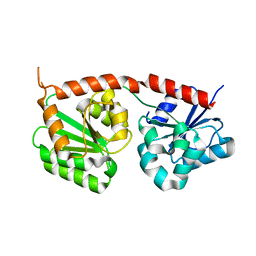 | | Se-glycosyltransferase RsSenB | | Descriptor: | Se glycosyltransferase | | Authors: | Huang, W, Long, F. | | Deposit date: | 2023-07-24 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of RsSenB at 2.15 Angstroms resolution
To Be Published
|
|
9ARV
 
 | | CryoEM structure of AMETA-A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, Isoform 1 of Immunoglobulin heavy constant mu | | Authors: | Huang, W, Sang, Z, Taylor, D. | | Deposit date: | 2024-02-23 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Adaptive multi-epitope targeting and avidity-enhanced nanobody platform for ultrapotent, durable antiviral therapy.
Cell, 2024
|
|
7UO0
 
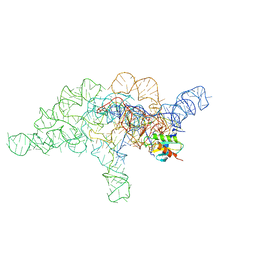 | | E.coli RNaseP Holoenzyme with Mg2+ | | Descriptor: | CALCIUM ION, Precursor tRNA substrate G(-1) G(-2), RNase P RNA, ... | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
7UO1
 
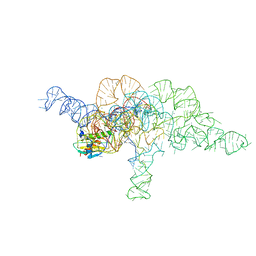 | | E.coli RNaseP Holoenzyme with Mg2+ | | Descriptor: | CALCIUM ION, E.coli RNase P RNA, Precursor tRNA substrate U(-1) and A(-2), ... | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
7UO2
 
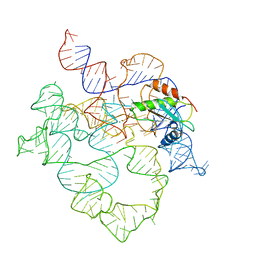 | | E.coli RNaseP Holoenzyme with Mg2+ | | Descriptor: | MAGNESIUM ION, RNase P RNA, Ribonuclease P protein component | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
7UO5
 
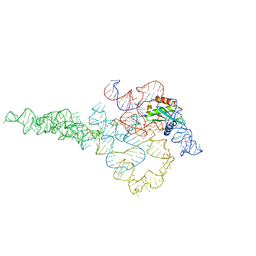 | | E.coli RNaseP Holoenzyme with Mg2+ | | Descriptor: | CALCIUM ION, RNase P RNA, Ribonuclease P protein component | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and mechanistic basis for recognition of alternative tRNA precursor substrates by bacterial ribonuclease P.
Nat Commun, 13, 2022
|
|
8IR4
 
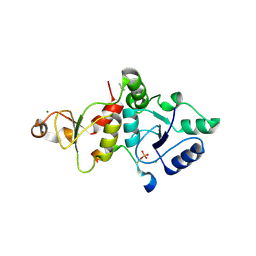 | | Crystal structure of the SLF1 BRCT domain in complex with a Rad18 peptide containing pS442 | | Descriptor: | CHLORIDE ION, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Xiang, S, Huang, W, Qiu, F. | | Deposit date: | 2023-03-17 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural insights into Rad18 targeting by the SLF1 BRCT domains.
J.Biol.Chem., 299, 2023
|
|
