4XOX
 
 | | Structure of beta-ketoacyl-ACP synthase I (FabB) from Vibrio Cholerae | | Descriptor: | 3-oxoacyl-ACP synthase | | Authors: | Hou, J, Grabowski, M, Cymborowski, M, Zheng, H, Cooper, D.R, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-16 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of beta-ketoacyl-ACP synthase I (FabB) from Vibrio Cholerae
to be published
|
|
6PNV
 
 | | 1.42 Angstrom Resolution Crystal Structure of Translocation Protein TolB from Salmonella enterica | | Descriptor: | POTASSIUM ION, SODIUM ION, Tol-Pal system protein TolB | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-03 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | 1.42 Angstrom Resolution Crystal Structure of Translocation Protein TolB from Salmonella enterica
To Be Published
|
|
5L09
 
 | | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica in complex with 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide | | Descriptor: | 1,2-ETHANEDIOL, 3-oxo-N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide, ACETIC ACID, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-26 | | Release date: | 2016-09-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Quorum-Sensing Transcriptional Activator from Yersinia enterocolitica
To Be Published
|
|
5KXJ
 
 | | Crystal Structure of L-Aspartate Oxidase from Salmonella typhimurium in the Complex with Substrate L-Aspartate | | Descriptor: | 1,2-ETHANEDIOL, ASPARTIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Osipiuk, J, Mulligan, R, Makowska-Grzyska, M, Maltseva, N, Shatsman, S, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-20 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of L-Aspartate Oxidase from Salmonella typhimurium in the Complex with Substrate L-Aspartate
To Be Published
|
|
5JPI
 
 | | 2.15 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with D-Eritadenine and NAD | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenosylhomocysteinase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-03 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with D-Eritadenine and NAD.
To Be Published
|
|
5JXW
 
 | | 2.25 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Neplanocin-A and NAD | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenosylhomocysteinase, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-13 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2.25 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Neplanocin-A and NAD
To Be Published
|
|
5KKO
 
 | | A 1.55A X-Ray Structure from Vibrio cholerae O1 biovar El Tor of a Hypothetical Protein | | Descriptor: | Uncharacterised protein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-22 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A 1.55A X-Ray Structure from Vibrio cholerae O1 biovar El Tor of a Hypothetical Protein
To Be Published
|
|
5KZ6
 
 | | 1.25 Angstrom Crystal Structure of Chitinase from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, Chitinase, SODIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-22 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.252 Å) | | Cite: | 1.25 Angstrom Crystal Structure of Chitinase from Bacillus anthracis.
To Be Published
|
|
5KWS
 
 | | Crystal Structure of Galactose Binding Protein from Yersinia pestis in the Complex with beta D Glucose | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-19 | | Release date: | 2016-08-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.316 Å) | | Cite: | Crystal Structure of Galactose Binding Protein from Yersinia pestis in the Complex with beta D Glucose
To Be Published
|
|
5K99
 
 | | Crystal structure of microcin immunity protein MccF from Bacillus anthracis in complex with McC | | Descriptor: | ADENOSINE MONOPHOSPHATE, Microcin C, Microcin C7 self-immunity protein mccF | | Authors: | Nocek, B, Kulikovsky, A, Severinov, K, Dubiley, S, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-31 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of microcin immunity protein MccF from Bacillus anthracis in complex with McC
To Be Published
|
|
5KJP
 
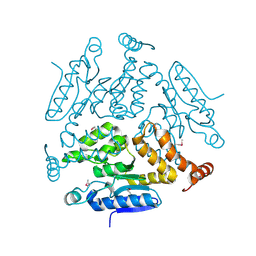 | |
5KVR
 
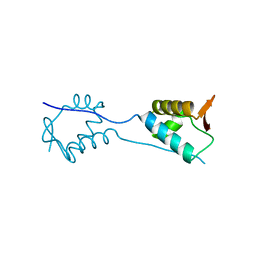 | | X-Ray Crystal Structure of a Fragment (1-75) of a Transcriptional Regulator PdhR from Escherichia coli CFT073 | | Descriptor: | Pyruvate dehydrogenase complex repressor | | Authors: | Brunzelle, J.S, Wawrzak, Z, Sandoval, J, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-15 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | X-Ray Crystal Structure of a Fragment (1-75) of a Transcriptional Regulator PdhR from Escherichia coli CFT073
To Be Published
|
|
6X9Y
 
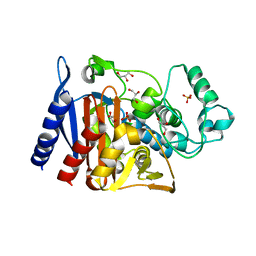 | | The crystal structure of a Beta-lactamase from Escherichia coli CFT073 | | Descriptor: | Beta-lactamase, GLYCEROL, S,R MESO-TARTARIC ACID, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a Beta-lactamase from Escherichia coli CFT073
To Be Published
|
|
6XS4
 
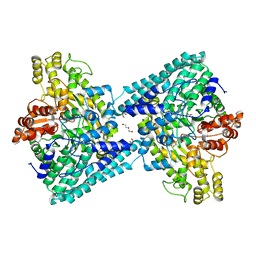 | | Crystal structure of glycyl radical enzyme ECL_02896 from Enterobacter cloacae subsp. cloacae | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Formate C-acetyltransferase | | Authors: | Valleau, D, Evdokimova, E, Stogios, P.J, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of glycyl radical enzyme ECL_02896 from Enterobacter cloacae subsp. cloacae.
To Be Published
|
|
6XJ3
 
 | | Crystal structure of Class D beta-lactamase from Klebsiella quasipneumoniae in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Class D beta-lactamase from Klebsiella quasipneumoniae
To Be Published
|
|
6WTC
 
 | | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2 | | Descriptor: | ACETIC ACID, Non-structural protein 7, Non-structural protein 8 | | Authors: | Wilamowski, M, Kim, Y, Jedrzejczak, R, Maltseva, N, Endres, M, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Second Form of the Co-factor Complex of NSP7 and the C-terminal Domain of NSP8 from SARS CoV-2
To Be Published
|
|
5HD6
 
 | | High resolution structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from Yersinia pestis at 1.35 A | | Descriptor: | 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase, GLYCEROL | | Authors: | Chang, C, Maltseva, N, Kim, Y, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-04 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High resolution structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from Yersinia pestis at 1.35 A
To Be Published
|
|
6MUK
 
 | | 1.93 Angstrom Resolution Crystal Structure of Peptidase M23 from Neisseria gonorrhoeae. | | Descriptor: | Peptidase M23, ZINC ION | | Authors: | Minasov, G, Shuvalova, L, Pshenychnyi, S, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-23 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | 1.93 Angstrom Resolution Crystal Structure of Peptidase M23 from Neisseria gonorrhoeae.
To Be Published
|
|
5I4K
 
 | | Metal ABC transporter from Listeria monocytogenes with manganese | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Manganese-binding lipoprotein MntA | | Authors: | Osipiuk, J, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-02-12 | | Release date: | 2016-02-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Metal ABC transporter from Listeria monocytogenes with manganese
to be published
|
|
6X2D
 
 | | Crystal Structure of DNase I Domain of Ribonuclease E from Vibrio cholerae | | Descriptor: | IODIDE ION, Ribonuclease E | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-20 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of DNase I Domain of Ribonuclease E from Vibrio cholerae.
To Be Published
|
|
6WY4
 
 | | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-12 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
5DIB
 
 | | 2.25 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) Y450L point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-08-31 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2.25 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) Y450L point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289
To Be Published
|
|
5DUL
 
 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis in complex with NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Osipiuk, J, Mulligan, R, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis in complex with NADPH .
to be published
|
|
6MGG
 
 | | Succinyl-CoA synthase from Francisella tularensis, phosphorylated, in complex with CoA | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Maltseva, N, Jedrzejczak, R, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-13 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Succinyl-CoA synthase from Francisella tularensis
to be published
|
|
5HW2
 
 | | Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with fumaric acid | | Descriptor: | 1,2-ETHANEDIOL, Adenylosuccinate lyase, FUMARIC ACID, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-28 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with fumaric acid
To Be Published
|
|
