479D
 
 | |
421D
 
 | |
419D
 
 | |
474D
 
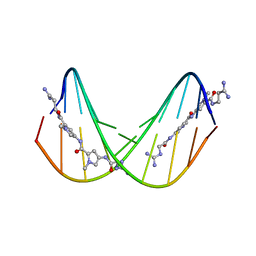 | | A NOVEL END-TO-END BINDING OF TWO NETROPSINS TO THE DNA DECAMER D(CCCCCIIIII)2 | | Descriptor: | DNA (5'-D(*CP*CP*CP*(CBR)P*CP*IP*IP*IP*IP*I)-3'), NETROPSIN | | Authors: | Chen, X, Rao, S.T, Sekar, K, Sundaralingam, M. | | Deposit date: | 1998-01-14 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel end-to-end binding of two netropsins to the DNA decamers d(CCCCCIIIII)2, d(CCCBr5CCIIIII)2and d(CBr5CCCCIIIII)2.
Nucleic Acids Res., 26, 1998
|
|
472D
 
 | | STRUCTURE OF AN OCTAMER RNA WITH TANDEM GG/UU MISPAIRS | | Descriptor: | RNA (5'-R(*GP*UP*AP*GP*GP*CP*AP*C)-3'), RNA (5'-R(*GP*UP*GP*UP*UP*UP*AP*C)-3') | | Authors: | Deng, J, Sundaralingam, M. | | Deposit date: | 1999-05-14 | | Release date: | 2000-11-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and crystal structure of an octamer RNA r(guguuuac)/r(guaggcac) with G.G/U.U tandem wobble base pairs: comparison with other tandem G.U pairs.
Nucleic Acids Res., 28, 2000
|
|
4ADL
 
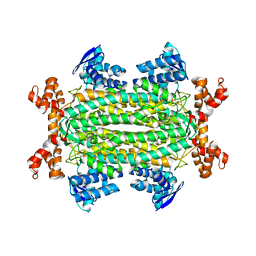 | | Crystal structures of Rv1098c in complex with malate | | Descriptor: | (2S)-2-hydroxybutanedioic acid, FUMARATE HYDRATASE CLASS II | | Authors: | Mechaly, A.E, Haouz, A, Miras, I, Weber, P, Shepard, W, Cole, S, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2011-12-26 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational Changes Upon Ligand Binding in the Essential Class II Fumarase Rv1098C from Mycobacterium Tuberculosis.
FEBS Lett., 586, 2012
|
|
4ADM
 
 | | Crystal structure of Rv1098c in complex with meso-tartrate | | Descriptor: | FUMARATE HYDRATASE CLASS II, GLYCEROL, S,R MESO-TARTARIC ACID | | Authors: | Mechaly, A.E, Haouz, A, Miras, I, Weber, P, Shepard, W, Cole, S, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2011-12-27 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Conformational Changes Upon Ligand Binding in the Essential Class II Fumarase Rv1098C from Mycobacterium Tuberculosis.
FEBS Lett., 586, 2012
|
|
100D
 
 | |
118D
 
 | |
159D
 
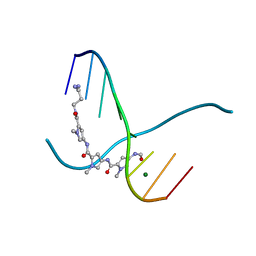 | | SIDE BY SIDE BINDING OF TWO DISTAMYCIN A DRUGS IN THE MINOR GROOVE OF AN ALTERNATING B-DNA DUPLEX | | Descriptor: | DISTAMYCIN A, DNA (5'-D(*IP*CP*IP*CP*IP*CP*IP*C)-3'), MAGNESIUM ION | | Authors: | Chen, X, Ramakrishnan, B, Rao, S.T, Sundaralingam, M. | | Deposit date: | 1994-02-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of two distamycin A molecules in the minor groove of an alternating B-DNA duplex.
Nat.Struct.Biol., 1, 1994
|
|
194D
 
 | | X-RAY STRUCTURES OF THE B-DNA DODECAMER D(CGCGTTAACGCG) WITH AN INVERTED CENTRAL TETRANUCLEOTIDE AND ITS NETROPSIN COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*TP*AP*AP*CP*GP*CP*G)-3') | | Authors: | Balendiran, K, Rao, S.T, Sekharudu, C.Y, Zon, G, Sundaralingam, M. | | Deposit date: | 1994-10-04 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structures of the B-DNA dodecamer d(CGCGTTAACGCG) with an inverted central tetranucleotide and its netropsin complex.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
2GRB
 
 | |
1CXW
 
 | | THE SECOND TYPE II MODULE FROM HUMAN MATRIX METALLOPROTEINASE 2 | | Descriptor: | HUMAN MATRIX METALLOPROTEINASE 2 | | Authors: | Briknarova, K, Grishaev, A, Banyai, L, Tordai, H, Patthy, L, Llinas, M. | | Deposit date: | 1999-08-31 | | Release date: | 1999-11-12 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | The second type II module from human matrix metalloproteinase 2: structure, function and dynamics.
Structure Fold.Des., 7, 1999
|
|
160D
 
 | |
161D
 
 | |
2VA1
 
 | | Crystal structure of UMP kinase from Ureaplasma parvum | | Descriptor: | PHOSPHATE ION, URIDYLATE KINASE | | Authors: | Egeblad-Welin, L, Welin, M, Wang, L, Eriksson, S. | | Deposit date: | 2007-08-28 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Investigations of Ureaplasma Parvum Ump Kinase - a Potential Antibacterial Drug Target
FEBS J., 274, 2007
|
|
195D
 
 | | X-RAY STRUCTURES OF THE B-DNA DODECAMER D(CGCGTTAACGCG) WITH AN INVERTED CENTRAL TETRANUCLEOTIDE AND ITS NETROPSIN COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*TP*AP*AP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Balendiran, K, Rao, S.T, Sekharudu, C.Y, Zon, G, Sundaralingam, M. | | Deposit date: | 1994-10-04 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structures of the B-DNA dodecamer d(CGCGTTAACGCG) with an inverted central tetranucleotide and its netropsin complex.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1DQH
 
 | |
1DQF
 
 | |
2L0S
 
 | | Solution Structure of Human Plasminogen Kringle 3 | | Descriptor: | Plasminogen | | Authors: | Christen, M.T, Frank, P, Schaller, J, Llinas, M. | | Deposit date: | 2010-07-15 | | Release date: | 2010-08-04 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Human plasminogen kringle 3: solution structure, functional insights, phylogenetic landscape.
Biochemistry, 49, 2010
|
|
2Y6K
 
 | | Xylotetraose bound to X-2 engineered mutated CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, CITRIC ACID, XYLANASE, ... | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
2G91
 
 | |
1DVL
 
 | |
2X31
 
 | | Modelling of the complex between subunits BchI and BchD of magnesium chelatase based on single-particle cryo-EM reconstruction at 7.5 ang | | Descriptor: | MAGNESIUM-CHELATASE 38 KDA SUBUNIT, MAGNESIUM-CHELATASE 60 KDA SUBUNIT | | Authors: | Lunqvist, J, Elmlund, H, Peterson Wulff, R, Berglund, L, Elmlund, D, Emanuelsson, C, Hebert, H, Willows, R.D, Hansson, M, Lindahl, M, Al-Karadaghi, S. | | Deposit date: | 2010-01-19 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | ATP-Induced Conformational Dynamics in the Aaa+ Motor Unit of Magnesium Chelatase.
Structure, 18, 2010
|
|
2VEN
 
 | | Structure-based enzyme engineering efforts with an inactive monomeric TIM variant: the importance of a single point mutation for generating an active site with suitable binding properties | | Descriptor: | CITRIC ACID, GLYCOSOMAL TRIOSEPHOSPHATE ISOMERASE | | Authors: | Alahuhta, M, Salin, M, Casteleijn, M.G, Kemmer, C, El-Sayed, I, Augustyns, K, Neubauer, P, Wierenga, R.K. | | Deposit date: | 2007-10-25 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Protein Engineering Efforts with a Monomeric Tim Variant: The Importance of a Single Point Mutation for Generating an Active Site with Suitable Binding Properties.
Protein Eng.Des.Sel., 21, 2008
|
|
