8CYE
 
 | |
8CWM
 
 | |
8SDB
 
 | | Crystal Structure of E.Coli Branching Enzyme in complex with malto-octose | | Descriptor: | 1,4-alpha-glucan branching enzyme GlgB, alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Bingham, C.R, Nayebi, H, Fawaz, R, Geiger, J.H. | | Deposit date: | 2023-04-06 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Maltooctaose-Bound Escherichia coli Branching Enzyme Suggests a Mechanism for Donor Chain Specificity.
Molecules, 28, 2023
|
|
8CXM
 
 | | Cryo-EM structure of the supercoiled E. coli K12 flagellar filament core, Normal waveform | | Descriptor: | Flagellin | | Authors: | Sonani, R.R, Kreutzberger, M.A.B, Sebastian, A.L, Scharf, B, Egelman, E.H. | | Deposit date: | 2022-05-21 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Convergent evolution in the supercoiling of prokaryotic flagellar filaments.
Cell, 185, 2022
|
|
8CVI
 
 | |
6CZV
 
 | | BRD4(BD1) complexed with 2759 | | Descriptor: | 1,2-ETHANEDIOL, 1-benzyl-5-(3,5-dimethyl-1,2-oxazol-4-yl)pyridin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Design and Characterization of Novel Covalent Bromodomain and Extra-Terminal Domain (BET) Inhibitors Targeting a Methionine.
J. Med. Chem., 61, 2018
|
|
6D5E
 
 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 1-[(2S)-1-{6-chloro-1-[(4-fluoro-3,5-dimethylphenyl)methyl]-2-(piperazin-1-yl)-1H-benzimidazol-4-yl}pyrrolidin-2-yl]methanamine, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
6D55
 
 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 6-chloro-2-(2,6-diazaspiro[3.3]heptan-2-yl)-1-[(4-fluoro-3,5-dimethylphenyl)methyl]-4-(4-methylpiperazin-1-yl)-1H-benzimidazole, FORMIC ACID, GLYCEROL, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
5VRL
 
 | | CRYSTAL STRUCTURE OF THE INHA FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH AN12855, EBSI 4333. | | Descriptor: | (~{N}~{E})-~{N}-[[2-[[2-ethylsulfonyl-1,1-bis(oxidanyl)-3,4-dihydro-2,3,1$l^{4}-benzodiazaborinin-7-yl]oxy]-5-(trifluoromethyl)phenyl]methylidene]hydroxylamine, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Abendroth, J, Edwards, T.E, Lorimer, D. | | Deposit date: | 2017-05-11 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a cofactor-independent inhibitor ofMycobacterium tuberculosisInhA.
Life Sci Alliance, 1, 2018
|
|
6D5G
 
 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 6-chloro-1-[(4-fluoro-3,5-dimethylphenyl)methyl]-2-(piperazin-1-yl)-4-(1,2,3,6-tetrahydropyridin-4-yl)-1H-benzimidazole, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
6D5V
 
 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 1-[(3-chloro-4-fluorophenyl)methyl]-5,6-dimethyl-1H-benzimidazol-2-amine, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
6D4B
 
 | | Crystal structure of Candida boidinii formate dehydrogenase V123A mutant complexed with NAD+ and azide | | Descriptor: | AZIDE ION, CHLORIDE ION, Formate dehydrogenase, ... | | Authors: | Guo, Q, Ye, H, Gakhar, L, Cheatum, C.M, Kohen, A. | | Deposit date: | 2018-04-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Oscillatory Active-site Motions Correlate with Kinetic Isotope Effects in Formate Dehydrogenase
Acs Catalysis, 2019
|
|
6D5W
 
 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 10-[(4-fluorophenyl)methyl]-2,3,4,10-tetrahydropyrimido[1,2-a]benzimidazole, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.478 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
6D56
 
 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 6-chloro-2-(2,6-diazaspiro[3.3]heptan-2-yl)-4-(3,5-dimethyl-1H-pyrazol-4-yl)-1-[(4-fluoro-3,5-dimethylphenyl)methyl]-1H-benzimidazole, FORMIC ACID, GLYCEROL, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
6D5J
 
 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 6-chloro-1-[(4-fluoro-3,5-dimethylphenyl)methyl]-2-(piperazin-1-yl)-1H-benzimidazole, FORMIC ACID, GLYCEROL, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
8GF2
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies eCR3022.20 and CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 Fab heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Broadening a SARS-CoV-1-neutralizing antibody for potent SARS-CoV-2 neutralization through directed evolution.
Sci.Signal., 16, 2023
|
|
3LN4
 
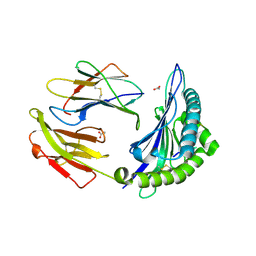 | | Crystal structure of HLA-B*4103 in complex with a 16mer self-peptide derived from heterogeneous nuclear ribonucleoproteins C1/C2 | | Descriptor: | 16-mer peptide from Heterogeneous nuclear ribonucleoproteins C1/C2, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Theodossis, A, Gras, S, Rossjohn, J. | | Deposit date: | 2010-02-01 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.296 Å) | | Cite: | The impact of human leukocyte antigen (HLA) micropolymorphism on ligand specificity within the HLA-B*41 allotypic family
Haematologica, 96, 2011
|
|
6D59
 
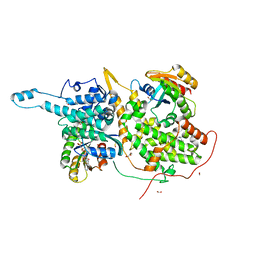 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 6-chloro-4-(3,5-dimethyl-1H-pyrazol-4-yl)-1-[(4-fluoro-3,5-dimethylphenyl)methyl]-2-(piperazin-1-yl)-1H-benzimidazole, FORMIC ACID, GLYCEROL, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
4BKQ
 
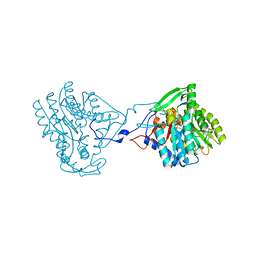 | | Enoyl-ACP reductase from Yersinia pestis (wildtype)with cofactor NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, PUTATIVE REDUCTASE YPZ3_3519 | | Authors: | Hirschbeck, M.W, Neckles, C, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-04-29 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia Pestis Fabv Enoyl-Acp Reductase.
Biochemistry, 55, 2016
|
|
6D4C
 
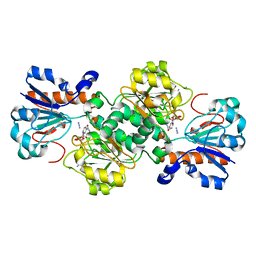 | | Crystal structure of Candida boidinii formate dehydrogenase V123G mutant complexed with NAD+ and azide | | Descriptor: | AZIDE ION, CHLORIDE ION, Formate dehydrogenase, ... | | Authors: | Guo, Q, Ye, H, Gakhar, L, Cheatum, C.M, Kohen, A. | | Deposit date: | 2018-04-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Oscillatory Active-site Motions Correlate with Kinetic Isotope Effects in Formate Dehydrogenase
Acs Catalysis, 2019
|
|
6D5M
 
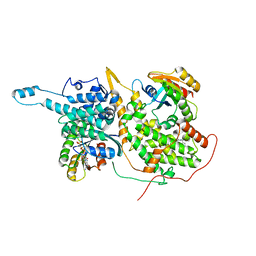 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 1-[(3-chloro-4-fluorophenyl)methyl]-5,6-dimethyl-2-(piperazin-1-yl)-1H-benzimidazole, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
6D5L
 
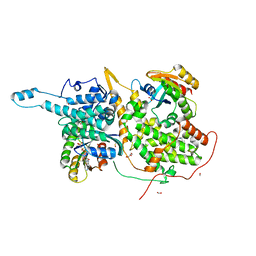 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 6-chloro-1-[(3-chloro-4-fluorophenyl)methyl]-2-(piperazin-1-yl)-1H-benzimidazole, FORMIC ACID, GLYCEROL, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
7QI2
 
 | |
3LN5
 
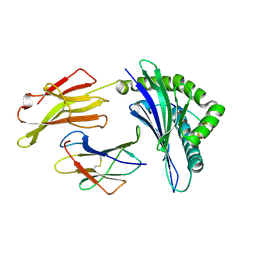 | | Crystal structure of HLA-B*4104 in complex with a 11mer self-peptide derived from S-methyl-5-thioadenosine phosphorylase | | Descriptor: | 11-mer peptide from S-methyl-5'-thioadenosine phosphorylase, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Theodossis, A, Gras, S, Rossjohn, J. | | Deposit date: | 2010-02-02 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The impact of human leukocyte antigen (HLA) micropolymorphism on ligand specificity within the HLA-B*41 allotypic family
Haematologica, 96, 2011
|
|
7M6T
 
 | | Crystal structure of SOCS2/ElonginB/ElonginC bound to a non-canonical peptide that enhances phospho-peptide binding | | Descriptor: | Elongin-B, Elongin-C, Non-canonical peptide F3, ... | | Authors: | Kershaw, N.J, Li, K, Linossi, E.M, Nicholson, S.E. | | Deposit date: | 2021-03-26 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Discovery of an exosite on the SOCS2-SH2 domain that enhances SH2 binding to phosphorylated ligands.
Nat Commun, 12, 2021
|
|
