6NPA
 
 | | X-ray crystal structure of TmpB, (R)-1-hydroxy-2-trimethylaminoethylphosphonate oxygenase, with (R)-1-hydroxy-2-trimethylaminoethylphosphonate | | Descriptor: | (2R)-2-hydroxy-N,N,N-trimethyl-2-phosphonoethan-1-aminium, FE (II) ION, FE (III) ION, ... | | Authors: | Rajakovich, L.J, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A New Microbial Pathway for Organophosphonate Degradation Catalyzed by Two Previously Misannotated Non-Heme-Iron Oxygenases.
Biochemistry, 58, 2019
|
|
6NPC
 
 | | X-ray crystal structure of TmpA, 2-trimethylaminoethylphosphonate hydroxylase, with Fe, 2OG, and 2-trimethylaminoethylphosphonate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, N,N,N-trimethyl-2-phosphonoethan-1-aminium, ... | | Authors: | Rajakovich, L.J, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A New Microbial Pathway for Organophosphonate Degradation Catalyzed by Two Previously Misannotated Non-Heme-Iron Oxygenases.
Biochemistry, 58, 2019
|
|
2MMA
 
 | | NMR-based docking model of GrxS14-BolA2 apo-heterodimer from Arabidopsis thaliana | | Descriptor: | BolA2, Monothiol glutaredoxin-S14, chloroplastic | | Authors: | Roret, T, Tsan, P, Couturier, J, Rouhier, N, Didierjean, C. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and Spectroscopic Insights into BolA-Glutaredoxin Complexes.
J.Biol.Chem., 289, 2014
|
|
2MM9
 
 | | Solution structure of reduced BolA2 from Arabidopsis thaliana | | Descriptor: | BolA2 | | Authors: | Roret, T, Tsan, P, Couturier, J, Rouhier, N, Didierjean, C. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and Spectroscopic Insights into BolA-Glutaredoxin Complexes.
J.Biol.Chem., 289, 2014
|
|
7CYR
 
 | |
6MTQ
 
 | | Crystal structure of VRC42.N1 Fab in complex with T117-F MPER scaffold | | Descriptor: | Antibody VRC42.N1 Fab heavy chain, Antibody VRC42.N1 Fab light chain, VRC42 epitope T117-F scaffold | | Authors: | Kwon, Y.D, Law, W.H, Veradi, R, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
6NPB
 
 | | X-ray crystal structure of TmpA, 2-trimethylaminoethylphosphonate hydroxylase, with Fe and 2OG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, SULFATE ION, ... | | Authors: | Rajakovich, L.J, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A New Microbial Pathway for Organophosphonate Degradation Catalyzed by Two Previously Misannotated Non-Heme-Iron Oxygenases.
Biochemistry, 58, 2019
|
|
6NTV
 
 | | SFTSV L endonuclease domain | | Descriptor: | RNA polymerase | | Authors: | Wang, W, Amarasinghe, G.K. | | Deposit date: | 2019-01-30 | | Release date: | 2020-01-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Cap-Snatching SFTSV Endonuclease Domain Is an Antiviral Target.
Cell Rep, 30, 2020
|
|
6NZ7
 
 | |
6NPD
 
 | | X-ray crystal structure of TmpA, 2-trimethylaminoethylphosphonate hydroxylase, with Fe, 2OG, and (R)-1-hydroxy-2-trimethylaminoethylphosphonate | | Descriptor: | (2R)-2-hydroxy-N,N,N-trimethyl-2-phosphonoethan-1-aminium, FE (II) ION, TmpA, ... | | Authors: | Rajakovich, L.J, Mitchell, A.J, Boal, A.K. | | Deposit date: | 2019-01-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A New Microbial Pathway for Organophosphonate Degradation Catalyzed by Two Previously Misannotated Non-Heme-Iron Oxygenases.
Biochemistry, 58, 2019
|
|
3U1S
 
 | | Crystal structure of human Fab PGT145, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | Fab PGT145 Heavy chain, Fab PGT145 Light chain, GLYCEROL, ... | | Authors: | Julien, J.-P, Diwanji, D, Burton, D.R, Wilson, I.A. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
5V7J
 
 | | Crystal Structure at 3.7 A Resolution of Glycosylated HIV-1 Clade A BG505 SOSIP.664 Prefusion Env Trimer with Four Glycans (N197, N276, N362, and N462) removed in Complex with Neutralizing Antibodies 3H+109L and 35O22. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 35O22 Fab heavy chain, ... | | Authors: | Stewart-Jones, G.B.E, Zhou, T, Kwong, P.D. | | Deposit date: | 2017-03-20 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Quantification of the Impact of the HIV-1-Glycan Shield on Antibody Elicitation.
Cell Rep, 19, 2017
|
|
6J72
 
 | | Crystal structure of IniA from Mycobacterium smegmatis with GTP bound | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Isoniazid inducible gene protein IniA, L(+)-TARTARIC ACID, ... | | Authors: | Wang, M.F, Guo, X.Y, Hu, J.J, Li, J, Rao, Z.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mycobacterial dynamin-like protein IniA mediates membrane fission.
Nat Commun, 10, 2019
|
|
5V9T
 
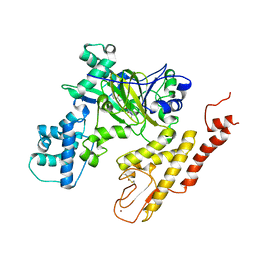 | | Crystal structure of selective pyrrolidine amide KDM5a inhibitor N-{(3R)-1-[3-(propan-2-yl)-1H-pyrazole-5-carbonyl]pyrrolidin-3-yl}cyclopropanecarboxamide (compound 48) | | Descriptor: | Lysine-specific demethylase 5A, N-{(3R)-1-[3-(propan-2-yl)-1H-pyrazole-5-carbonyl]pyrrolidin-3-yl}cyclopropanecarboxamide, NICKEL (II) ION, ... | | Authors: | Kiefer, J.R, Liang, J, Vinogradova, M. | | Deposit date: | 2017-03-23 | | Release date: | 2017-05-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | From a novel HTS hit to potent, selective, and orally bioavailable KDM5 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
3V65
 
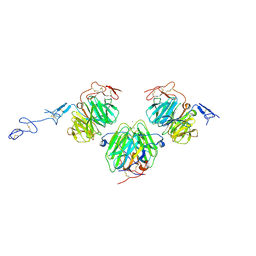 | | Crystal structure of agrin and LRP4 complex | | Descriptor: | Agrin, CALCIUM ION, Low-density lipoprotein receptor-related protein 4 | | Authors: | Zong, Y, Jin, R. | | Deposit date: | 2011-12-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of agrin-LRP4-MuSK signaling.
Genes Dev., 26, 2012
|
|
7VAH
 
 | |
7U9O
 
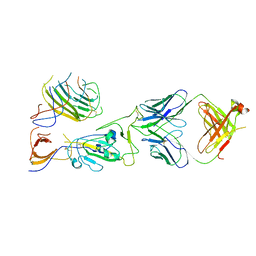 | | SARS-CoV-2 spike trimer RBD in complex with Fab NE12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NE12 Fab heavy chain, NE12 Fab light chain, ... | | Authors: | Tsybovsky, Y, Kwong, P.D, Farci, P. | | Deposit date: | 2022-03-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Potent monoclonal antibodies neutralize Omicron sublineages and other SARS-CoV-2 variants.
Cell Rep, 41, 2022
|
|
7U9P
 
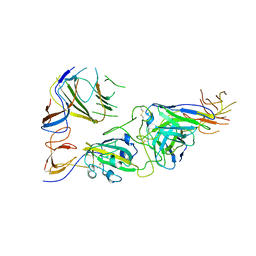 | | SARS-CoV-2 spike trimer RBD in complex with Fab NA8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NA8 Fab heavy chain, NA8 Fab light chain, ... | | Authors: | Tsybovsky, Y, Kwong, P.D, Farci, P. | | Deposit date: | 2022-03-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Potent monoclonal antibodies neutralize Omicron sublineages and other SARS-CoV-2 variants.
Cell Rep, 41, 2022
|
|
8V13
 
 | | Crystal structure of Black mamba toxin in complex with Centi-SNX-B03 antibody | | Descriptor: | Centi-SNX-B03 Fab heavy chain, Centi-SNX-B03 Fab light chain, SODIUM ION, ... | | Authors: | Pletnev, S, Glanville, J, Kwong, P.D. | | Deposit date: | 2023-11-19 | | Release date: | 2025-01-29 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Snake venom protection by a cocktail of varespladib and broadly neutralizing human antibodies.
Cell, 188, 2025
|
|
5WAL
 
 | |
5WEV
 
 | |
3U4E
 
 | | Crystal Structure of PG9 Fab in Complex with V1V2 Region from HIV-1 strain CAP45 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PG9 Heavy Chain, PG9 Light Chain, ... | | Authors: | Gorman, J, McLellan, J, Pancera, M, Kwong, P.D. | | Deposit date: | 2011-10-07 | | Release date: | 2011-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3U2S
 
 | | Crystal Structure of PG9 Fab in Complex with V1V2 Region from HIV-1 strain ZM109 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | McLellan, J.S, Pancera, M, Kwong, P.D. | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9.
Nature, 480, 2011
|
|
3UDD
 
 | | Tankyrase-1 in complex with small molecule inhibitor | | Descriptor: | 3-(4-methoxyphenyl)-5-({[4-(4-methoxyphenyl)-5-methyl-4H-1,2,4-triazol-3-yl]sulfanyl}methyl)-1,2,4-oxadiazole, GLYCEROL, SULFATE ION, ... | | Authors: | Kirby, C.A, Stams, T. | | Deposit date: | 2011-10-28 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | [1,2,4]triazol-3-ylsulfanylmethyl)-3-phenyl-[1,2,4]oxadiazoles: antagonists of the Wnt pathway that inhibit tankyrases 1 and 2 via novel adenosine pocket binding.
J.Med.Chem., 55, 2012
|
|
8X0S
 
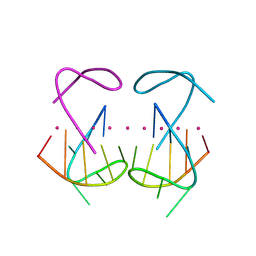 | | Crystal structure of r(G4C2)2 | | Descriptor: | POTASSIUM ION, RNA (5'-R(*GP*GP*GP*GP*CP*CP*GP*GP*GP*GP*C)-3') | | Authors: | Geng, Y, Liu, C, Cai, Q, Zhu, G. | | Deposit date: | 2023-11-05 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.956 Å) | | Cite: | Crystal structure of a tetrameric RNA G-quadruplex formed by hexanucleotide repeat expansions of C9orf72 in ALS/FTD.
Nucleic Acids Res., 52, 2024
|
|
