7PNQ
 
 | |
4PQ6
 
 | |
4PQB
 
 | |
5MF1
 
 | |
4PQC
 
 | |
7ESX
 
 | | Crystal structure of Wolbachia cytoplasmic incompatibility factor CidA from wPip | | Descriptor: | Bacteria factor 1 | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic insights into the complexes formed by Wolbachia cytoplasmic incompatibility factors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ESY
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidA and CidBND1-ND2 from wPip | | Descriptor: | Bacteria factor 1, CALCIUM ION, ULP_PROTEASE domain-containing protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Structural and mechanistic insights into the complexes formed by Wolbachia cytoplasmic incompatibility factors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ESZ
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CinA and CinB with Mn2+ from wPip | | Descriptor: | BACTERIA FACTOR A, BACTERIA FACTOR B, MANGANESE (II) ION | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.476 Å) | | Cite: | Structural and mechanistic insights into the complexes formed by Wolbachia cytoplasmic incompatibility factors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ET0
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CinA and CinB from wPip | | Descriptor: | Bacteria factor A, Bacteria factor B | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-05-12 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic insights into the complexes formed by Wolbachia cytoplasmic incompatibility factors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3DOR
 
 | | Crystal Structure of mature CPAF | | Descriptor: | Protein CT_858, SULFATE ION | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-07-06 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3DPM
 
 | | Structure of mature CPAF complexed with lactacystin | | Descriptor: | N-acetyl-S-({(2R,3S,4R)-3-hydroxy-2-[(1S)-1-hydroxy-2-methylpropyl]-4-methyl-5-oxopyrrolidin-2-yl}carbonyl)cysteine, Protein CT_858 | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-07-09 | | Release date: | 2009-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3DJA
 
 | | Crystal Structure of cpaf solved with MAD | | Descriptor: | Protein CT_858 | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-06-22 | | Release date: | 2009-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3DPN
 
 | | Crystal Structure of cpaf s499a mutant | | Descriptor: | Protein CT_858 | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-07-09 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
7Y5K
 
 | | Crystal structure of human CAF-1 core complex in spacegroup C2221 | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, GLYCEROL, ... | | Authors: | Liu, C.P, Wang, M.Z, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y61
 
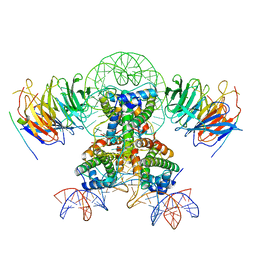 | | Cryo-EM structure of the two CAF1LCs bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5O
 
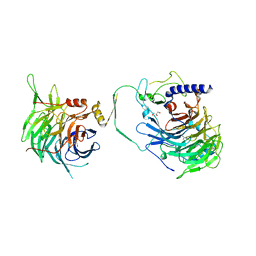 | | Crystal structure of human CAF-1 core complex in spacegroup P21 | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, GLYCEROL, ... | | Authors: | Liu, C.P, Wang, M.Z, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5V
 
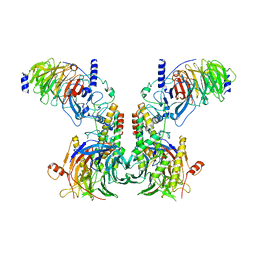 | | Cryo-EM structure of the dimeric human CAF1LC-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, C, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y60
 
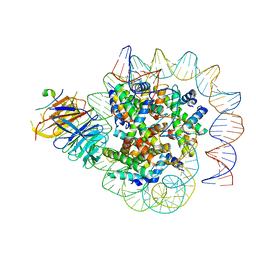 | | Cryo-EM structure of human CAF1LC bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, C, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2022-06-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5L
 
 | | Crystal structure of human CAF-1 core complex in spacegroup C2 | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, GLYCEROL, ... | | Authors: | Liu, C.P, Wang, M.Z, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5W
 
 | | Cryo-EM structure of the left-handed Di-tetrasome | | Descriptor: | Histone H3.1, Histone H4, Widom 601 DNA (147-MER) | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5U
 
 | | Cryo-EM structure of the monomeric human CAF1LC-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
3OG4
 
 | | The crystal structure of human interferon lambda 1 complexed with its high affinity receptor in space group P21212 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin 28 receptor, alpha (Interferon, ... | | Authors: | Miknis, Z.J, Magracheva, E, Lei, W, Zdanov, A, Kotenko, S.V, Wlodawer, A. | | Deposit date: | 2010-08-16 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of the complex of human interferon-lambda1 with its high affinity receptor interferon-lambdaR1.
J.Mol.Biol., 404, 2010
|
|
8HUR
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with S217622 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUW
 
 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with S217622 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Wang, J, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
8HUS
 
 | | Crystal structure of SARS main protease in complex with S217622 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2022-12-24 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
