3SB6
 
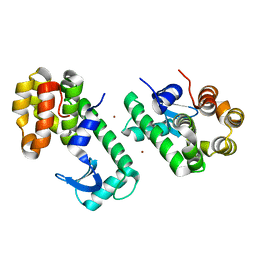 | | Cu-mediated Dimer of T4 Lysozyme D61H/K65H/R76H/R80H by Synthetic Symmetrization | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Lysozyme | | Authors: | Soriaga, A.B, Laganowsky, A, Zhao, M, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-03 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
1OIN
 
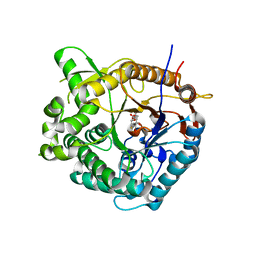 | | Family 1 b-glucosidase from Thermotoga maritima | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-glucopyranose, BETA-GLUCOSIDASE A | | Authors: | Gloster, T, Zechel, D.L, Boraston, A.B, Boraston, C.M, Macdonald, J.M, Tilbrook, D.M, Stick, R.V, Davies, G.J. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Iminosugar Glycosidase Inhibitors: Structural and Thermodynamic Dissection of the Binding of Isofagomine and 1-Deoxynojirimycin to Beta-Glucosidases
J.Am.Chem.Soc., 125, 2003
|
|
3S9L
 
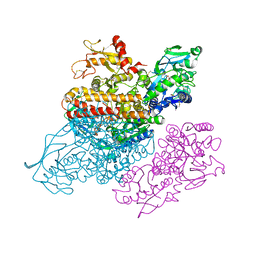 | | Complex between transferrin receptor 1 and transferrin with iron in the N-Lobe, cryocooled 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eckenroth, B.E, Steere, A.N, Mason, A.B, Everse, S.J. | | Deposit date: | 2011-06-01 | | Release date: | 2011-08-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | How the binding of human transferrin primes the transferrin receptor potentiating iron release at endosomal pH.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1OH4
 
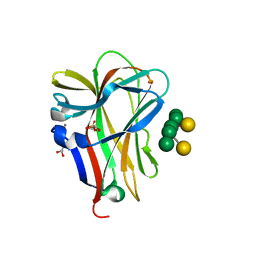 | | Structural and thermodynamic dissection of specific mannan recognition by a carbohydrate-binding module | | Descriptor: | BETA-MANNOSIDASE, CALCIUM ION, GLYCEROL, ... | | Authors: | Boraston, A.B, Revett, T.J, Boraston, C.M, Nurizzo, D, Davies, G.J. | | Deposit date: | 2003-05-21 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Thermodynamic Dissection of Specific Mannan Recognition by a Carbohydrate Binding Module, Tmcbm27
Structure, 11, 2003
|
|
3SEU
 
 | | Zn-mediated Polymer of Maltose-binding Protein A216H/K220H by Synthetic Symmetrization (Form III) | | Descriptor: | ACETATE ION, GLYCEROL, Maltose-binding periplasmic protein, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
3SEX
 
 | | Ni-mediated Dimer of Maltose-binding Protein A216H/K220H by Synthetic Symmetrization (Form II) | | Descriptor: | IODIDE ION, Maltose-binding periplasmic protein, NICKEL (II) ION, ... | | Authors: | Zhao, M, Soriaga, A.B, Laganowsky, A, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2011-06-11 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An approach to crystallizing proteins by metal-mediated synthetic symmetrization.
Protein Sci., 20, 2011
|
|
1OFN
 
 | | Purification, crystallisation and preliminary structural studies of dTDP-4-keto-6-deoxy-glucose-5-epimerase (EvaD) from Amycolatopsis orientalis; the fourth enzyme in the dTDP-L-epivancosamine biosynthetic pathway. | | Descriptor: | GLYCEROL, PCZA361.16 | | Authors: | Merkel, A.B, Naismith, J.H. | | Deposit date: | 2003-04-17 | | Release date: | 2004-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Purification, Crystallization and Preliminary Structural Studies of Dtdp-4-Keto-6-Deoxy-Glucose-5-Epimerase (Evad) from Amycolatopsis Orientalis, the Fourth Enzyme in the Dtdp-L-Epivancosamine Biosynthetic Pathway.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1NPV
 
 | | Crystal structure of HIV-1 protease complexed with LDC271 | | Descriptor: | POL polyprotein, {1-BENZYL-3-[2-BENZYL-3-OXO-4-(1-OXO-1,2,3,4-TETRAHYDRO- ISOQUINOLIN-4-YL)-2,3-DIHYDRO-1H-PYRROL-2-YL]-2- HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Smith, A.B. | | Deposit date: | 2003-01-20 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis, and biological evaluation of monopyrrolinone-based HIV-1 protease inhibitors.
J.Med.Chem., 46, 2003
|
|
1QZQ
 
 | | human Tyrosyl DNA phosphodiesterase | | Descriptor: | tyrosyl-DNA phosphodiesterase 1 | | Authors: | Raymond, A.C, Rideout, M.C, Staker, B, Hjerrild, K, Burgin Jr, A.B. | | Deposit date: | 2003-09-17 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Analysis of Human Tyrosyl-DNA Phosphodiesterase I Catalytic Residues.
J.Mol.Biol., 338, 2004
|
|
4TLW
 
 | | CARDS TOXIN, FULL-LENGTH | | Descriptor: | ADP-ribosylating toxin CARDS | | Authors: | Becker, A, GALALELDEEN, A, Taylor, A.B, Hart, P.J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of CARDS toxin, a unique ADP-ribosylating and vacuolating cytotoxin from Mycoplasma pneumoniae.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7JNB
 
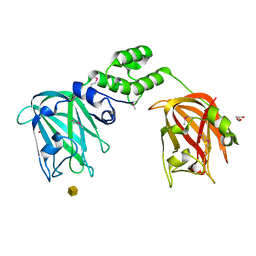 | |
7JS4
 
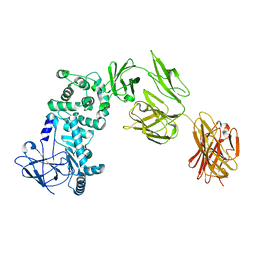 | |
1Q0Y
 
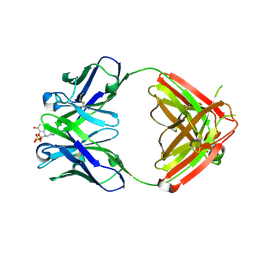 | | Anti-Morphine Antibody 9B1 Complexed with Morphine | | Descriptor: | (7R,7AS,12BS)-3-METHYL-2,3,4,4A,7,7A-HEXAHYDRO-1H-4,12-METHANO[1]BENZOFURO[3,2-E]ISOQUINOLINE-7,9-DIOL, Fab 9B1, Heavy chain, ... | | Authors: | Pozharski, E, Wilson, M.A, Hewagama, A, Shanafelt, A.B, Petsko, G, Ringe, D. | | Deposit date: | 2003-07-17 | | Release date: | 2004-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Anchoring a cationic ligand: the structure of the Fab fragment of the anti-morphine antibody 9B1 and its complex with morphine
J.Mol.Biol., 337, 2004
|
|
4PEF
 
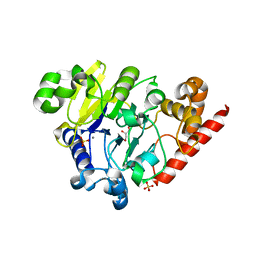 | | Dbr1 in complex with sulfate | | Descriptor: | GLYCEROL, MANGANESE (II) ION, RNA lariat debranching enzyme, ... | | Authors: | Montemayor, E.J, Katolik, A, Clark, N.E, Taylor, A.B, Schuermann, J.P, Combs, D.J, Johnsson, R, Holloway, S.P, Stevens, S.W, Damha, M.J, Hart, P.J. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis of lariat RNA recognition by the intron debranching enzyme Dbr1.
Nucleic Acids Res., 42, 2014
|
|
7JWF
 
 | | Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2020-08-25 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | The structure of a family 110 glycoside hydrolase provides insight into the hydrolysis of alpha-1,3-galactosidic linkages in lambda-carrageenan and blood group antigens.
J.Biol.Chem., 295, 2020
|
|
3QT9
 
 | | Analysis of a new family of widely distributed metal-independent alpha mannosidases provides unique insight into the processing of N-linked glycans, Clostridium perfringens CPE0426 complexed with alpha-1,6-linked 1-thio-alpha-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein CPE0426, alpha-D-mannopyranose-(1-6)-6-thio-alpha-D-mannopyranose | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
7JRM
 
 | |
1QW0
 
 | | Crystal Structure of Haemophilus influenzae N175L mutant Holo Ferric ion-Binding Protein A | | Descriptor: | FE (III) ION, Iron-utilization periplasmic protein, PHOSPHATE ION | | Authors: | Shouldice, S.R, Skene, R.J, Dougan, D.R, McRee, D.E, Tari, L.W, Schryvers, A.B. | | Deposit date: | 2003-08-29 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Presence of ferric hydroxide clusters in mutants of Haemophilus influenzae ferric ion-binding protein A
Biochemistry, 42, 2003
|
|
1QVS
 
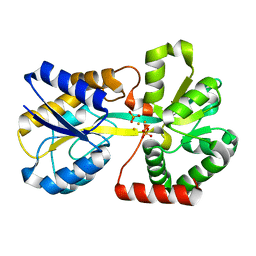 | | Crystal Structure of Haemophilus influenzae H9A mutant Holo Ferric ion-Binding Protein A | | Descriptor: | FE (III) ION, Iron-utilization periplasmic protein, PHOSPHATE ION | | Authors: | Shouldice, S.R, Skene, R.J, Dougan, D.R, McRee, D.E, Tari, L.W, Schryvers, A.B. | | Deposit date: | 2003-08-28 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Presence of ferric hydroxide clusters in mutants of Haemophilus influenzae ferric ion-binding protein A
Biochemistry, 42, 2003
|
|
7JND
 
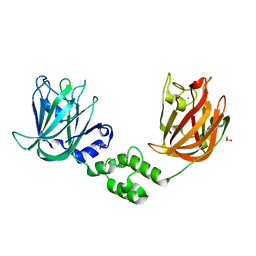 | |
7JNF
 
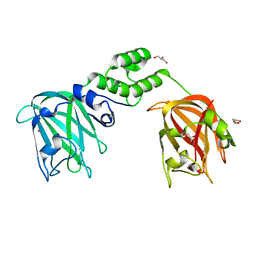 | |
3QT3
 
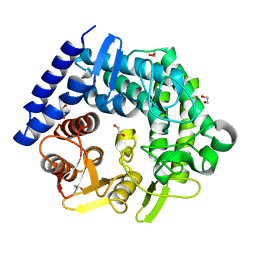 | | Analysis of a New Family of Widely Distributed Metal-independent alpha-Mannosidases Provides Unique Insight into the Processing of N-linked Glycans, Clostridium perfringens CPE0426 apo-structure | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein CPE0426 | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
4TKP
 
 | | Complex of Ubc13 with the RING domain of the TRIM5alpha retroviral restriction factor | | Descriptor: | SULFATE ION, Tripartite motif-containing protein 5, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Johnson, R, Taylor, A.B, Hart, P.J, Ivanov, D.N. | | Deposit date: | 2014-05-27 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | RING Dimerization Links Higher-Order Assembly of TRIM5 alpha to Synthesis of K63-Linked Polyubiquitin.
Cell Rep, 12, 2015
|
|
4TLV
 
 | | CARDS TOXIN, NICKED | | Descriptor: | ACETATE ION, ADP-ribosylating toxin CARDS, GLYCEROL, ... | | Authors: | Taylor, A.B, Pakhomova, O.N, Hart, P.J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of CARDS toxin, a unique ADP-ribosylating and vacuolating cytotoxin from Mycoplasma pneumoniae.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7JFS
 
 | |
