3WMZ
 
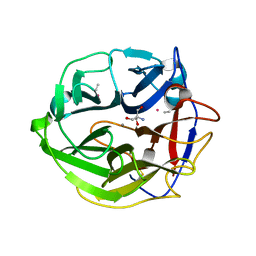 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase ethylmercury derivative | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ETHYL MERCURY ION, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3WD3
 
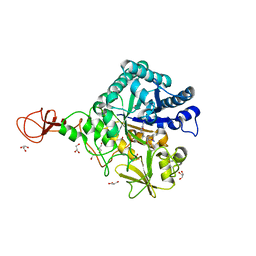 | | Serratia marcescens Chitinase B complexed with azide inhibitor | | Descriptor: | Chitinase B, GLYCEROL, SULFATE ION, ... | | Authors: | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WN1
 
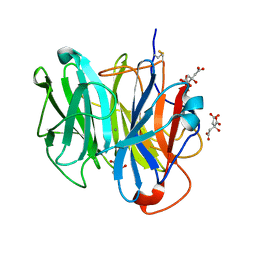 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase in complex with xylotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3WR7
 
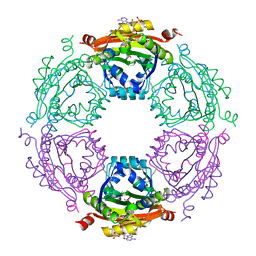 | | Crystal Structure of Spermidine Acetyltransferase from Escherichia coli | | Descriptor: | COENZYME A, SPERMIDINE, Spermidine N1-acetyltransferase | | Authors: | Sugiyama, S, Ishikawa, S, Tomitori, S, Niiyama, M, Hirose, M, Miyazaki, Y, Higashi, K, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Kashiwagi, K, Igarashi, K, Matsumura, H. | | Deposit date: | 2014-02-20 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism underlying promiscuous polyamine recognition by spermidine acetyltransferase
Int.J.Biochem.Cell Biol., 76, 2016
|
|
3WD0
 
 | | Serratia marcescens Chitinase B, tetragonal form | | Descriptor: | Chitinase B, DITHIANE DIOL, GLYCEROL, ... | | Authors: | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WN0
 
 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase in complex with L-arabinose | | Descriptor: | CALCIUM ION, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
7NJJ
 
 | | Proteinase K grown inside HARE serial crystallography chip | | Descriptor: | NITRATE ION, Proteinase K | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NKF
 
 | | Hen egg white lysozyme (HEWL) Grown inside (Not centrifuged) HARE serial crystallography chip. | | Descriptor: | Lysozyme, SODIUM ION | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-17 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJG
 
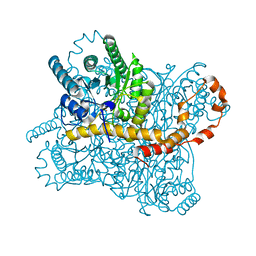 | | Xylose isomerase grown inside HARE serial crystallography chip | | Descriptor: | COBALT (II) ION, Xylose isomerase | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJI
 
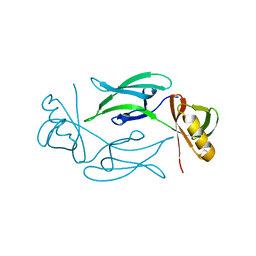 | | HEX1 (in cellulo) loaded on HARE serial crystallography chip | | Descriptor: | Woronin body major protein | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJE
 
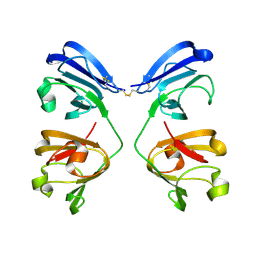 | | gamma(S)-crystallin 9-site deamidation mutant grown inside HARE serial crystallography chip | | Descriptor: | Gamma-crystallin S | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJH
 
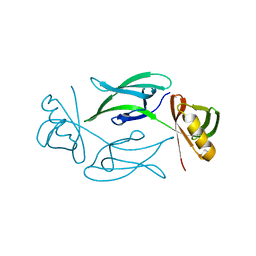 | | HEX1 (in cellulo) grown inside HARE serial crystallography chip | | Descriptor: | Woronin body major protein | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7NJF
 
 | | Hen egg white lysozyme (HEWL) grown inside HARE serial crystallography chip | | Descriptor: | Lysozyme, SODIUM ION | | Authors: | Norton-Baker, B, Mehrabi, P, Boger, J, Schonherr, R, von Stetten, D, Schikora, H, Martin, R.W, Miller, R.J.D, Redecke, L, Schulz, E.C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A simple vapor-diffusion method enables protein crystallization inside the HARE serial crystallography chip.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3VUS
 
 | | Escherichia coli PgaB N-terminal domain | | Descriptor: | ACETATE ION, MERCURY (II) ION, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase, ... | | Authors: | Nishiyama, T, Noguchi, H, Yoshida, H, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2012-07-05 | | Release date: | 2012-11-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of the deacetylase domain of Escherichia coli PgaB, an enzyme required for biofilm formation: a circularly permuted member of the carbohydrate esterase 4 family
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VSO
 
 | | Human PPAR gamma ligand binding domain in complex with a gamma selective agonist mekt21 | | Descriptor: | (2R)-2-benzyl-3-[4-propoxy-3-({[4-(pyrimidin-2-yl)benzoyl]amino}methyl)phenyl]propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Oyama, T, Waku, T, Ohashi, M, Morikawa, K, Miyachi, H. | | Deposit date: | 2012-04-30 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of a series of alpha-benzyl phenylpropanoic acid-type peroxisome proliferator-activated receptor (PPAR) gamma partial agonists with improved aqueous solubility
Bioorg.Med.Chem., 21, 2013
|
|
3WO4
 
 | | Crystal structure of the IL-18 signaling ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
3WO2
 
 | | Crystal structure of human interleukin-18 | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Interleukin-18, SULFATE ION | | Authors: | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
3WKJ
 
 | | The nucleosome containing human TSH2B | | Descriptor: | CHLORIDE ION, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Urahama, T, Horikoshi, N, Osakabe, A, Tachiwana, H, Kurumizaka, H. | | Deposit date: | 2013-10-22 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human nucleosome containing the testis-specific histone variant TSH2B.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3WO3
 
 | | Crystal structure of IL-18 in complex with IL-18 receptor alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
4BKM
 
 | | Crystal structure of the murine AUM (phosphoglycolate phosphatase) capping domain as a fusion protein with the catalytic core domain of murine chronophin (pyridoxal phosphate phosphatase) | | Descriptor: | MAGNESIUM ION, NITRATE ION, PYRIDOXAL PHOSPHATE PHOSPHATASE, ... | | Authors: | Knobloch, G, Seifried, A, Gohla, A, Schindelin, H. | | Deposit date: | 2013-04-26 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Evolutionary and Structural Analyses of the Mammalian Haloacid Dehalogenase-Type Phosphatases Aum and Chronophin Provide Insight Into the Basis of Their Different Substrate Specificities.
J.Biol.Chem., 289, 2014
|
|
4BX2
 
 | | Crystal Structure of murine Chronophin (Pyridoxal Phosphate Phosphatase) in complex with Beryllium trifluoride | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Knobloch, G, Gohla, A, Schindelin, H. | | Deposit date: | 2013-07-08 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Chronophin Dimerization is Required for Proper Positioning of its Substrate Specificity Loop.
J.Biol.Chem., 289, 2014
|
|
4BX3
 
 | |
4BX0
 
 | | Crystal Structure of a Monomeric Variant of murine Chronophin (Pyridoxal Phosphate phosphatase) | | Descriptor: | GLYCEROL, MAGNESIUM ION, PYRIDOXAL PHOSPHATE PHOSPHATASE | | Authors: | Kestler, C, Knobloch, G, Gohla, A, Schindelin, H. | | Deposit date: | 2013-07-08 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Chronophin Dimerization is Required for Proper Positioning of its Substrate Specificity Loop
J.Biol.Chem., 289, 2014
|
|
2ZQZ
 
 | | R-state structure of allosteric L-lactate dehydrogenase from Lactobacillus casei | | Descriptor: | L-lactate dehydrogenase, SULFATE ION | | Authors: | Arai, K, Ishimitsu, T, Fushinobu, S, Uchikoba, H, Matsuzawa, H, Taguchi, H. | | Deposit date: | 2008-08-22 | | Release date: | 2009-09-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active and inactive state structures of unliganded Lactobacillus casei allosteric L-lactate dehydrogenase.
Proteins, 78, 2010
|
|
2ZQY
 
 | | T-state structure of allosteric L-lactate dehydrogenase from Lactobacillus casei | | Descriptor: | L-lactate dehydrogenase, NITRATE ION | | Authors: | Arai, K, Ishimitsu, T, Fushinobu, S, Uchikoba, H, Matsuzawa, H, Taguchi, H. | | Deposit date: | 2008-08-22 | | Release date: | 2009-09-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Active and inactive state structures of unliganded Lactobacillus casei allosteric L-lactate dehydrogenase.
Proteins, 78, 2010
|
|
