9CTQ
 
 | | Best1 + GABA open state | | Descriptor: | Bestrophin-1, CALCIUM ION, GAMMA-AMINO-BUTANOIC ACID | | Authors: | Owji, A.P, Kittredge, A, Zhang, Y, Yang, T. | | Deposit date: | 2024-07-25 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | GAD65 tunes the functions of Best1 as a GABA receptor and a neurotransmitter conducting channel.
Nat Commun, 15, 2024
|
|
9CTS
 
 | | Best1 + GABA intermediate state 2 | | Descriptor: | Bestrophin-1, CALCIUM ION, GAMMA-AMINO-BUTANOIC ACID | | Authors: | Owji, A.P, Kittredge, A, Zhang, Y, Yang, T. | | Deposit date: | 2024-07-25 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | GAD65 tunes the functions of Best1 as a GABA receptor and a neurotransmitter conducting channel.
Nat Commun, 15, 2024
|
|
9CTT
 
 | | Best1 + GABA closed state | | Descriptor: | Bestrophin-1, CALCIUM ION | | Authors: | Owji, A.P, Kittredge, A, Zhang, Y, Yang, T. | | Deposit date: | 2024-07-25 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | GAD65 tunes the functions of Best1 as a GABA receptor and a neurotransmitter conducting channel.
Nat Commun, 15, 2024
|
|
6DUP
 
 | | CRYSTAL STRUCTURE OF PXR IN COMPLEX WITH COMPOUND 7 | | Descriptor: | (2S)-2-({[3'-(trifluoromethyl)[1,1'-biphenyl]-4-yl]oxy}methyl)-2,3-dihydro-7H-[1,3]oxazolo[3,2-a]pyrimidin-7-one, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Chen, X, Zhang, Y, Mclean, L.R. | | Deposit date: | 2018-06-21 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Amelioration of PXR-mediated CYP3A4 induction by mGluR2 modulators.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6KOP
 
 | | Mycobacterium tuberculosis initial transcription complex comprising sigma H and 5'-OH RNA of 9 nt | | Descriptor: | DNA (5'-D(*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*GP*TP*G)-3'), DNA (5'-D(*TP*TP*GP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*A)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Li, L, Zhang, Y. | | Deposit date: | 2019-08-12 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KOQ
 
 | | Mycobacterium tuberculosis initial transcription complex comprising sigma H and 5'-OH RNA of 10 nt | | Descriptor: | DNA (5'-D(*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*GP*TP*G)-3'), DNA (5'-D(*TP*TP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*A)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Li, L, Zhang, Y. | | Deposit date: | 2019-08-12 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.353 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KON
 
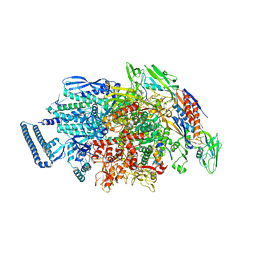 | | Mycobacterium tuberculosis initial transcription complex comprising sigma H and 5'-OH RNA of 5 nt | | Descriptor: | DNA (5'-D(P*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*GP*TP*G)-3'), DNA (5'-D(P*GP*TP*TP*GP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Li, L, Zhang, Y. | | Deposit date: | 2019-08-12 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KOO
 
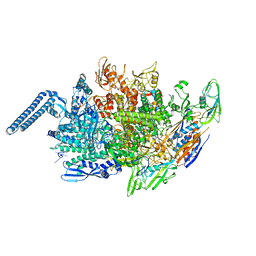 | | Mycobacterium tuberculosis initial transcription complex comprising sigma H and 5'-OH RNA of 7 nt | | Descriptor: | DNA (5'-D(*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*GP*TP*G)-3'), DNA (5'-D(*TP*TP*GP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*A)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Li, L, Zhang, Y. | | Deposit date: | 2019-08-12 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4IMJ
 
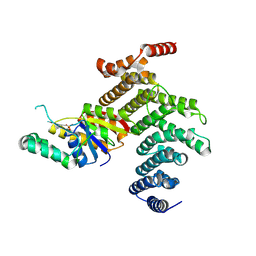 | | Novel Modifications on C-terminal Domain of RNA Polymerase II can Fine-tune the Phosphatase Activity of Ssu72 | | Descriptor: | CG14216, CTD, PHOSPHATE ION, ... | | Authors: | Luo, Y, Yogesha, S.D, Zhang, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-08-07 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Novel Modifications on C-terminal Domain of RNA Polymerase II Can Fine-tune the Phosphatase Activity of Ssu72.
Acs Chem.Biol., 8, 2013
|
|
1RLG
 
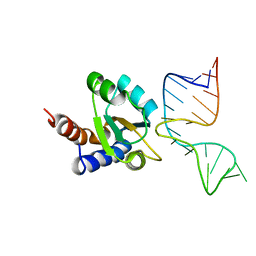 | | Molecular basis of Box C/D RNA-protein interaction: co-crystal structure of the Archaeal sRNP intiation complex | | Descriptor: | 25-MER, 50S ribosomal protein L7Ae | | Authors: | Moore, T, Zhang, Y, Fenley, M.O, Li, H. | | Deposit date: | 2003-11-25 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Basis of Box C/D RNA-Protein Interactions; Cocrystal Structure of Archaeal L7Ae and a Box C/D RNA.
STRUCTURE, 12, 2004
|
|
1RQI
 
 | | Active Conformation of Farnesyl Pyrophosphate Synthase Bound to Isopentyl Pyrophosphate and Dimethylallyl S-Thiolodiphosphate | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, DIPHOSPHATE, Geranyltranstransferase, ... | | Authors: | Hosfield, D.J, Zhang, Y, Dougan, D.R, Brooun, A, Tari, L.W, Swanson, R.V, Finn, J. | | Deposit date: | 2003-12-05 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis
J.Biol.Chem., 279, 2004
|
|
3QH2
 
 | | Crystal structure of TenI from Bacillus subtilis complexed with product cThz-P | | Descriptor: | 4-methyl-5-[2-(phosphonooxy)ethyl]-1,3-thiazole-2-carboxylic acid, Regulatory protein tenI, SULFATE ION | | Authors: | Han, Y, Zhang, Y, Hazra, A, Chatterjee, A, Lai, R, Begley, T.P, Ealick, S.E. | | Deposit date: | 2011-01-25 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | A Missing Enzyme in Thiamin Thiazole Biosynthesis: Identification of TenI as a Thiazole Tautomerase.
J.Am.Chem.Soc., 133, 2011
|
|
3ORW
 
 | | Crystal structure of thermophilic phosphotriesterase from Geobacillus kaustophilus HTA426 | | Descriptor: | COBALT (II) ION, Phosphotriesterase | | Authors: | Zheng, B.S, Yu, S.S, Zhang, Y, Lou, Z.Y, Feng, Y. | | Deposit date: | 2010-09-07 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of thermophilic phosphotriesterase from Geobacillus kaustophilus HTA426
To be Published
|
|
1SPX
 
 | | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form | | Descriptor: | short-chain reductase family member (5L265) | | Authors: | Schormann, N, Zhou, J, McCombs, D, Bray, T, Symersky, J, Huang, W.-Y, Luan, C.-H, Gray, R, Luo, D, Arabashi, A, Bunzel, B, Nagy, L, Lu, S, Li, S, Lin, G, Zhang, Y, Qiu, S, Tsao, J, Luo, M, Carson, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-17 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Glucose Dehydrogenase of Caenorhabditis Elegans in the Apo-Form: A Member of the SDR-Family
To be Published
|
|
1P32
 
 | | CRYSTAL STRUCTURE OF HUMAN P32, A DOUGHNUT-SHAPED ACIDIC MITOCHONDRIAL MATRIX PROTEIN | | Descriptor: | MITOCHONDRIAL MATRIX PROTEIN, SF2P32 | | Authors: | Jiang, J, Zhang, Y, Krainer, A.R, Xu, R.-M. | | Deposit date: | 1998-11-02 | | Release date: | 1999-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4IMI
 
 | | Novel Modifications on C-terminal Domain of RNA Polymerase II can Fine- tune the Phosphatase Activity of Ssu72. | | Descriptor: | CG14216, CTD, PHOSPHATE ION, ... | | Authors: | Luo, Y, Yogesha, S.D, Zhang, Y. | | Deposit date: | 2013-01-03 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel Modifications on C-terminal Domain of RNA Polymerase II Can Fine-tune the Phosphatase Activity of Ssu72.
Acs Chem.Biol., 8, 2013
|
|
1PFB
 
 | | Structural Basis for specific binding of polycomb chromodomain to histone H3 methylated at K27 | | Descriptor: | ACETIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Min, J.R, Zhang, Y, Xu, R.-M. | | Deposit date: | 2003-05-24 | | Release date: | 2003-10-07 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27.
Genes Dev., 17, 2003
|
|
1T7S
 
 | | Structural Genomics of Caenorhabditis elegans: Structure of BAG-1 protein | | Descriptor: | BAG-1 cochaperone | | Authors: | Symersky, J, Zhang, Y, Schormann, N, Li, S, Bunzel, R, Pruett, P, Luan, C.-H, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-10 | | Release date: | 2004-05-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: structure of the BAG domain.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SZP
 
 | | A Crystal Structure of the Rad51 Filament | | Descriptor: | DNA repair protein RAD51, SULFATE ION | | Authors: | Conway, A.B, Lynch, T.W, Zhang, Y, Fortin, G.S, Symington, L.S, Rice, P.A. | | Deposit date: | 2004-04-06 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of a Rad51 filament.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1RTR
 
 | | Crystal Structure of S. Aureus Farnesyl Pyrophosphate Synthase | | Descriptor: | geranyltranstransferase | | Authors: | Hosfield, D.J, Zhang, Y, Dougan, D.R, Brooun, A, Tari, L.W, Swanson, R.V, Finn, J. | | Deposit date: | 2003-12-10 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis
J.Mol.Biol., 279, 2004
|
|
1RQJ
 
 | | Active Conformation of Farnesyl Pyrophosphate Synthase Bound to Isopentyl Pyrophosphate and Risedronate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranyltranstransferase, ... | | Authors: | Hosfield, D.J, Zhang, Y, Dougan, D.R, Brooun, A, Tari, L.W, Swanson, R.V, Finn, J. | | Deposit date: | 2003-12-05 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis
J.Biol.Chem., 279, 2004
|
|
1T9F
 
 | | Structural genomics of Caenorhabditis elegans: Structure of a protein with unknown function | | Descriptor: | MALONATE ION, protein 1d10 | | Authors: | Symersky, J, Li, S, Bunzel, R, Schormann, N, Luo, D, Huang, W.Y, Qiu, S, Gray, R, Zhang, Y, Arabashi, A, Lu, S, Luan, C.H, Tsao, J, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-16 | | Release date: | 2004-05-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: Structure of a protein with unknown function.
To be Published
|
|
7LJC
 
 | | Allosteric modulator LY3154207 binding to SKF-81297-bound dopamine receptor 1 in complex with miniGs protein | | Descriptor: | (1R)-6-chloro-1-phenyl-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, ... | | Authors: | Zhuang, Y, Krumm, B, Zhang, H, Zhou, X.E, Wang, Y, Guo, J, Huang, X.-P, Liu, Y, Wang, L, Cheng, X, Jiang, Y, Jiang, H, Melcher, K, Zhang, C, Yi, W, Roth, B.L, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-03 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of dopamine binding and allosteric modulation of the human D1 dopamine receptor.
Cell Res., 31, 2021
|
|
7LJD
 
 | | Allosteric modulator LY3154207 binding to dopamine-bound dopamine receptor 1 in complex with miniGs protein | | Descriptor: | 2-[2,6-bis(chloranyl)phenyl]-1-[(1S,3R)-3-(hydroxymethyl)-1-methyl-5-(3-methyl-3-oxidanyl-butyl)-3,4-dihydro-1H-isoquinolin-2-yl]ethanone, CHOLESTEROL, D(1A) dopamine receptor, ... | | Authors: | Zhuang, Y, Krumm, B, Zhang, H, Zhou, X.E, Wang, Y, Guo, J, Huang, X.-P, Liu, Y, Wang, L, Cheng, X, Jiang, Y, Jiang, H, Melcher, K, Zhang, C, Yi, W, Roth, B.L, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-01-28 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of dopamine binding and allosteric modulation of the human D1 dopamine receptor.
Cell Res., 31, 2021
|
|
4N2C
 
 | | Crystal structure of Protein Arginine Deiminase 2 (F221/222A, 10 mM Ca2+) | | Descriptor: | CALCIUM ION, Protein-arginine deiminase type-2 | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.022 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
