7GCW
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with NAU-LAT-445f63e5-6 (Mpro-x10800) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-benzyloxan-4-yl)-N'-(pyridin-3-yl)urea | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GD0
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with BAR-COM-0f94fc3d-23 (Mpro-x10834) | | Descriptor: | (2S)-2-(3-bromophenyl)-2-hydroxy-N-(4-methoxypyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GG7
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with CHO-MSK-00c5269a-2 (Mpro-x12177) | | Descriptor: | 2-(1H-benzotriazol-1-yl)-N-[(3-chlorophenyl)methyl]-N-(4-methoxyphenyl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GGM
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-afd4d4fd-2 (Mpro-x12677) | | Descriptor: | 2-(6-chloropyridin-2-yl)-N-(isoquinolin-4-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GDB
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-968e8d9c-1 (Mpro-x10906) | | Descriptor: | (4S)-6-chloro-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5XX9
 
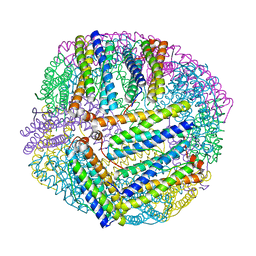 | |
4Z6K
 
 | |
8OE6
 
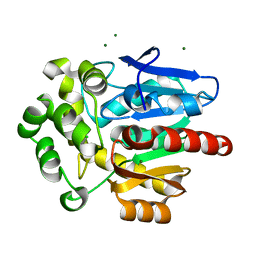 | | Structure of hyperstable haloalkane dehalogenase variant DhaA231 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Structure of hyperstable haloalkane dehalogenase variant DhaA231 | | Authors: | Marek, M. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Advancing Enzyme's Stability and Catalytic Efficiency through Synergy of Force-Field Calculations, Evolutionary Analysis, and Machine Learning.
Acs Catalysis, 13, 2023
|
|
8OE2
 
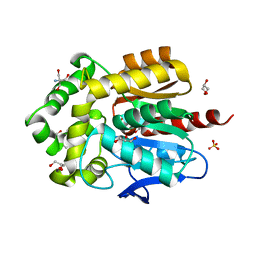 | | Structure of hyperstable haloalkane dehalogenase variant DhaA223 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Marek, M. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Advancing Enzyme's Stability and Catalytic Efficiency through Synergy of Force-Field Calculations, Evolutionary Analysis, and Machine Learning.
Acs Catalysis, 13, 2023
|
|
3N2J
 
 | | Azurin H117G, oxidized form | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | Hoffmann, M, Alagaratnam, S, Canters, G.W, Einsle, O. | | Deposit date: | 2010-05-18 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Probing the reactivity of different forms of azurin by flavin photoreduction.
Febs J., 278, 2011
|
|
7FCF
 
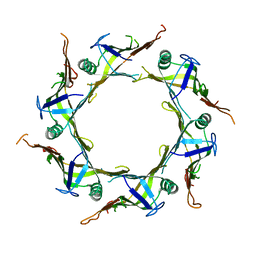 | | Crystal structure of T6SS Hcp protein | | Descriptor: | Fimbrial protein | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Bacterial antagonism of Chromobacterium haemolyticum and characterization of its putative type VI secretion system.
Res.Microbiol., 173, 2022
|
|
6S6E
 
 | |
6TY7
 
 | | Crystal structure of haloalkane dehalogenase variant DhaA115 domain-swapped dimer type-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Markova, K, Chaloupkova, R, Damborsky, J, Marek, M. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Computational Enzyme Stabilization Can Affect Folding Energy Landscapes and Lead to Catalytically Enhanced Domain-Swapped Dimers
Acs Catalysis, 11, 2021
|
|
1DMF
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF CALLINECTES SAPIDUS METALLOTHIONEIN-I DETERMINED BY HOMONUCLEAR AND HETERONUCLEAR MAGNETIC RESONANCE SPECTOSCOPY | | Descriptor: | CADMIUM ION, CD6 METALLOTHIONEIN-1 | | Authors: | Narula, S.S, Brouwer, M, Hua, Y, Armitage, I.M. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Callinectes sapidus metallothionein-1 determined by homonuclear and heteronuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
1DME
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF CALLINECTES SAPIDUS METALLOTHIONEIN-I DETERMINED BY HOMONUCLEAR AND HETERONUCLEAR MAGNETIC RESONANCE SPECTOSCOPY | | Descriptor: | CADMIUM ION, CD6 METALLOTHIONEIN-1 | | Authors: | Narula, S.S, Brouwer, M, Hua, Y, Armitage, I.M. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Callinectes sapidus metallothionein-1 determined by homonuclear and heteronuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
1DMC
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF CALLINECTES SAPIDUS METALLOTHIONEIN-I DETERMINED BY HOMONUCLEAR AND HETERONUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD6 METALLOTHIONEIN-1 | | Authors: | Narula, S.S, Brouwer, M, Hua, Y, Armitage, I.M. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Callinectes sapidus metallothionein-1 determined by homonuclear and heteronuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
1DMD
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF CALLINECTES SAPIDUS METALLOTHIONEIN-I DETERMINED BY HOMONUCLEAR AND HETERONUCLEAR MAGNETIC RESONANCE SPECTOSCOPY | | Descriptor: | CADMIUM ION, CD6 METALLOTHIONEIN-1 | | Authors: | Narula, S.S, Brouwer, M, Hua, Y, Armitage, I.M. | | Deposit date: | 1994-11-22 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of Callinectes sapidus metallothionein-1 determined by homonuclear and heteronuclear magnetic resonance spectroscopy.
Biochemistry, 34, 1995
|
|
6S97
 
 | |
6VCC
 
 | | Cryo-EM structure of the Dvl2 DIX filament | | Descriptor: | Segment polarity protein dishevelled homolog DVL-2 | | Authors: | Enos, M, Kan, W, Muennich, S, Chen, D.H, Skiniotis, G, Weis, W.I. | | Deposit date: | 2019-12-20 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Limited Dishevelled/Axin oligomerization determines efficiency of Wnt/ beta-catenin signal transduction.
Elife, 9, 2020
|
|
3PZE
 
 | | JNK1 in complex with inhibitor | | Descriptor: | 3-(carbamoylamino)-5-phenylthiophene-2-carboxamide, Mitogen-activated protein kinase 8, SULFATE ION | | Authors: | Xue, Y. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Checkpoint Kinase Inhibitor (S)-5-(3-Fluorophenyl)-N-(piperidin-3-yl)-3-ureidothiophene-2-carboxamide (AZD7762) by Structure-Based Design and Optimization of Thiophenecarboxamide Ureas.
J.Med.Chem., 55, 2012
|
|
6YN2
 
 | |
8JAX
 
 | |
8JB0
 
 | |
3ESZ
 
 | | K2AK3A Flavodoxin from Anabaena | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, GLYCEROL, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J, Goni, G, Medina, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase
Biochim.Biophys.Acta, 1787, 2009
|
|
3NZ0
 
 | | Non-phosphorylated TYK2 kinase with CMP6 | | Descriptor: | 2-TERT-BUTYL-9-FLUORO-3,6-DIHYDRO-7H-BENZ[H]-IMIDAZ[4,5-F]ISOQUINOLINE-7-ONE, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2010-07-15 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new regulatory switch in a JAK protein kinase.
Proteins, 79, 2011
|
|
