3WQ8
 
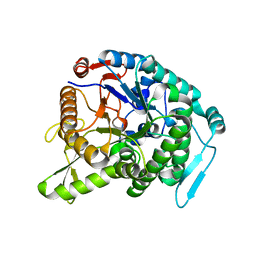 | | Monomer structure of hyperthermophilic beta-glucosidase mutant forming a dodecameric structure in the crystal form | | Descriptor: | Beta-glucosidase | | Authors: | Nakabayashi, M, Kataoka, M, Watanabe, M, Ishikawa, K. | | Deposit date: | 2014-01-23 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Monomer structure of a hyperthermophilic beta-glucosidase mutant forming a dodecameric structure in the crystal form.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3VHX
 
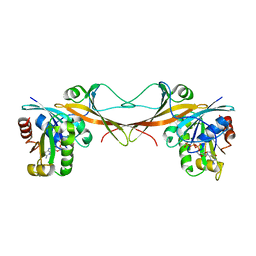 | | The crystal structure of Arf6-MKLP1 (Mitotic kinesin-like protein 1) complex | | Descriptor: | ADP-ribosylation factor 6, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Makyio, H, Takei, T, Ohgi, H, Takahashi, S, Takatsu, H, Ueda, T, Kanaho, Y, Xie, Y, Shin, H.W, Kamikubo, H, Kataoka, M, Kawasaki, M, Kato, R, Wakatsuki, S, Nakayama, K. | | Deposit date: | 2011-09-12 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for Arf6-MKLP1 complex formation on the Flemming body responsible for cytokinesis
Embo J., 31, 2012
|
|
3WDQ
 
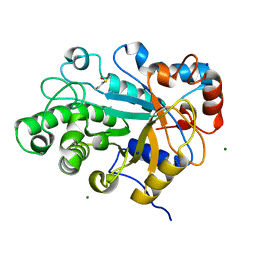 | | Crystal structure of beta-mannanase from a symbiotic protist of the termite Reticulitermes speratus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-mannanase, MAGNESIUM ION, ... | | Authors: | Tsukagoshi, H, Ishida, T, Touhara, K.K, Igarashi, K, Samejima, M, Fushinobu, S, Kitamoto, K, Arioka, M. | | Deposit date: | 2013-06-20 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and Biochemical Analyses of Glycoside Hydrolase Family 26 beta-Mannanase from a Symbiotic Protist of the Termite Reticulitermes speratus
J.Biol.Chem., 289, 2014
|
|
3WG8
 
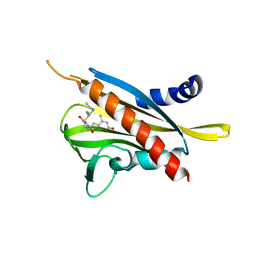 | | Crystal structure of the abscisic acid receptor PYR1 in complex with an antagonist AS6 | | Descriptor: | (2Z,4E)-5-[(1S)-3-(hexylsulfanyl)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Akiyama, T, Sue, M, Takeuchi, J, Okamoto, M, Muto, T, Endo, A, Nambara, E, Hirai, N, Ohnishi, T, Cutler, S.R, Todoroki, Y, Yajima, S. | | Deposit date: | 2013-07-31 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Designed abscisic acid analogs as antagonists of PYL-PP2C receptor interactions
Nat.Chem.Biol., 10, 2014
|
|
3UES
 
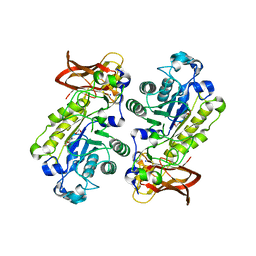 | | Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis complexed with deoxyfuconojirimycin | | Descriptor: | (2S,3R,4S,5R)-2-METHYLPIPERIDINE-3,4,5-TRIOL, 1,2-ETHANEDIOL, Alpha-1,3/4-fucosidase, ... | | Authors: | Sakurama, H, Fushinobu, S, Yoshida, E, Honda, Y, Hidaka, M, Ashida, H, Kitaoka, M, Katayama, T, Yamamoto, K, Kumagai, H. | | Deposit date: | 2011-10-31 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1,3-1,4-alpha-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains
J.Biol.Chem., 287, 2012
|
|
3UET
 
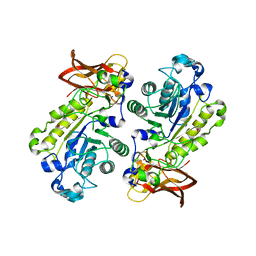 | | Crystal structure of alpha-1,3/4-fucosidase from Bifidobacterium longum subsp. infantis D172A/E217A mutant complexed with lacto-N-fucopentaose II | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,3/4-fucosidase, SODIUM ION, ... | | Authors: | Sakurama, H, Fushinobu, S, Yoshida, E, Honda, Y, Hidaka, M, Ashida, H, Kitaoka, M, Katayama, T, Yamamoto, K, Kumagai, H. | | Deposit date: | 2011-10-31 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1,3-1,4-alpha-L-fucosynthase that specifically introduces Lewis a/x antigens into type-1/2 chains
J.Biol.Chem., 287, 2012
|
|
3W7W
 
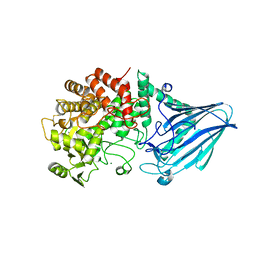 | | Crystal structure of E. coli YgjK E727A complexed with 2-O-alpha-D-glucopyranosyl-alpha-D-galactopyranose | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Uncharacterized protein YgjK, ... | | Authors: | Miyazaki, T, Ichikawa, M, Yokoi, G, Kitaoka, M, Mori, H, Kitano, Y, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2013-03-08 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a bacterial glycoside hydrolase family 63 enzyme in complex with its glycosynthase product, and insights into the substrate specificity.
Febs J., 280, 2013
|
|
3WDP
 
 | | Structural analysis of a beta-glucosidase mutant derived from a hyperthermophilic tetrameric structure | | Descriptor: | Beta-glucosidase, GLYCEROL, PHOSPHATE ION | | Authors: | Nakabayashi, M, Kataoka, M, Ishikawa, K. | | Deposit date: | 2013-06-19 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of beta-glucosidase mutants derived from a hyperthermophilic tetrameric structure.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WI7
 
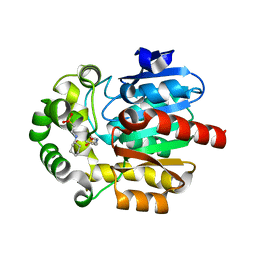 | | Crystal Structure of the Novel Haloalkane Dehalogenase DatA from Agrobacterium tumefaciens C58 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Haloalkane dehalogenase | | Authors: | Guan, L.J, Yabuki, H, Okai, M, Ohtsuka, J, Tanokura, M. | | Deposit date: | 2013-09-06 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the novel haloalkane dehalogenase DatA from Agrobacterium tumefaciens C58 reveals a special halide-stabilizing pair and enantioselectivity mechanism.
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3WIB
 
 | | Crystal structure of Y109W Mutant Haloalkane Dehalogenase DatA from Agrobacterium tumefaciens C58 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Haloalkane dehalogenase | | Authors: | Guan, L.J, Yabuki, H, Okai, M, Ohtsuka, J, Tanokura, M. | | Deposit date: | 2013-09-09 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the novel haloalkane dehalogenase DatA from Agrobacterium tumefaciens C58 reveals a special halide-stabilizing pair and enantioselectivity mechanism.
Appl.Microbiol.Biotechnol., 98, 2014
|
|
3W7X
 
 | | Crystal structure of E. coli YgjK D324N complexed with melibiose | | Descriptor: | CALCIUM ION, Uncharacterized protein YgjK, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose | | Authors: | Miyazaki, T, Ichikawa, M, Yokoi, G, Kitaoka, M, Mori, H, Kitano, Y, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2013-03-08 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a bacterial glycoside hydrolase family 63 enzyme in complex with its glycosynthase product, and insights into the substrate specificity.
Febs J., 280, 2013
|
|
3WWH
 
 | | Crystal structure of the first R-stereoselective -transaminase identified from Arthrobacter sp. KNK168 (FERM-BP-5228) | | Descriptor: | (R)-amine transaminase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Guan, L.J, Ohtsuka, J, Okai, M, Miyakawa, T, Mase, T, Zhi, Y, Ito, N, Yasohara, Y, Tanokura, M. | | Deposit date: | 2014-06-18 | | Release date: | 2015-08-12 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A new target region for changing the substrate specificity of amine transaminases.
Sci Rep, 5, 2015
|
|
3WUI
 
 | | Dimeric horse cytochrome c formed by refolding from molten globule state | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Deshpande, M.S, Parui, P.P, Kamikubo, H, Yamanaka, M, Nagao, S, Komori, H, Kataoka, M, Higuchi, Y, Hirota, S. | | Deposit date: | 2014-04-25 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Formation of domain-swapped oligomer of cytochrome C from its molten globule state oligomer.
Biochemistry, 53, 2014
|
|
3WWJ
 
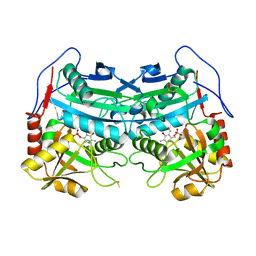 | | Crystal structure of an engineered sitagliptin-producing transaminase, ATA-117-Rd11 | | Descriptor: | (R)-amine transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Guan, L.J, Ohtsuka, J, Okai, M, Miyakawa, T, Mase, T, Zhi, Y, Hou, F, Ito, N, Yasohara, Y, Tanokura, M. | | Deposit date: | 2014-06-18 | | Release date: | 2015-08-12 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A new target region for changing the substrate specificity of amine transaminases.
Sci Rep, 5, 2015
|
|
3X3Y
 
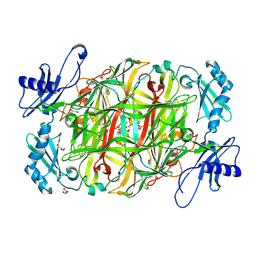 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by histamine | | Descriptor: | COPPER (II) ION, GLYCEROL, POTASSIUM ION, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X42
 
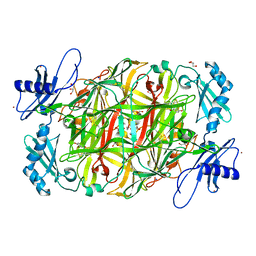 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis in the presence of sodium bromide | | Descriptor: | BROMIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | Deposit date: | 2015-03-10 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.875 Å) | | Cite: | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3VSC
 
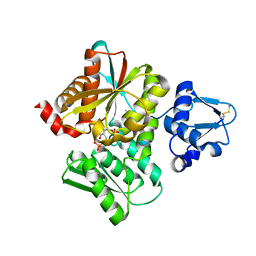 | | Crystal Structure of the K127A Mutant of O-Phosphoserine Sulfhydrylase Complexed with External Schiff Base of Pyridoxal 5'-Phosphate with O-Phospho-L-Serine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHOSERINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nakamura, T, Kawai, Y, Kataoka, M, Ishikawa, K. | | Deposit date: | 2012-04-24 | | Release date: | 2012-05-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural analysis of the substrate recognition mechanism in O-phosphoserine sulfhydrylase from the hyperthermophilic archaeon Aeropyrum pernix K1
J.Mol.Biol., 422, 2012
|
|
3VSA
 
 | | Crystal Structure of O-phosphoserine sulfhydrylase without acetate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PYRIDOXAL-5'-PHOSPHATE, Protein CysO | | Authors: | Nakamura, T, Kawai, Y, Kataoka, M, Ishikawa, K. | | Deposit date: | 2012-04-24 | | Release date: | 2012-05-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural analysis of the substrate recognition mechanism in O-phosphoserine sulfhydrylase from the hyperthermophilic archaeon Aeropyrum pernix K1
J.Mol.Biol., 422, 2012
|
|
3VSD
 
 | | Crystal Structure of the K127A Mutant of O-Phosphoserine Sulfhydrylase Complexed with External Schiff Base of Pyridoxal 5'-Phosphate with O-Acetyl-L-Serine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, O-ACETYLSERINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nakamura, T, Kawai, Y, Kataoka, M, Ishikawa, K. | | Deposit date: | 2012-04-24 | | Release date: | 2012-05-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural analysis of the substrate recognition mechanism in O-phosphoserine sulfhydrylase from the hyperthermophilic archaeon Aeropyrum pernix K1
J.Mol.Biol., 422, 2012
|
|
3VVG
 
 | |
3VKM
 
 | | Protease-resistant mutant form of Human Galectin-8 in complex with sialyllactose and lactose | | Descriptor: | 1,2-ETHANEDIOL, Galectin-8, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Yoshida, H, Yamashita, S, Teraoka, M, Nakakita, S, Nishi, N, Kamitori, S. | | Deposit date: | 2011-11-18 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | X-ray structure of a protease-resistant mutant form of human galectin-8 with two carbohydrate recognition domains
Febs J., 279, 2012
|
|
3WJL
 
 | | Crystal structure of IIb selective Fc variant, Fc(V12), in complex with FcgRIIb | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, Low affinity immunoglobulin gamma Fc region receptor II-c, ... | | Authors: | Kadono, S, Mimoto, F, Katada, H, Igawa, T, Kuramochi, T, Muraoka, M, Wada, Y, Haraya, K, Miyazaki, T, Hattori, K. | | Deposit date: | 2013-10-11 | | Release date: | 2013-11-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Engineered antibody Fc variant with selectively enhanced Fc gamma RIIb binding over both Fc gamma RIIaR131 and Fc gamma RIIaH131.
Protein Eng.Des.Sel., 26, 2013
|
|
3WC8
 
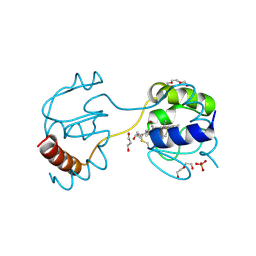 | | Dimeric horse cytochrome c obtained by refolding with desalting method | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Parui, P.P, Deshpande, M.S, Nagao, S, Kamikubo, H, Komori, H, Higuchi, Y, Kataoka, M, Hirota, S. | | Deposit date: | 2013-05-25 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Formation of Oligomeric Cytochrome c during Folding by Intermolecular Hydrophobic Interaction between N- and C-Terminal alpha-Helices
Biochemistry, 52, 2013
|
|
3WJJ
 
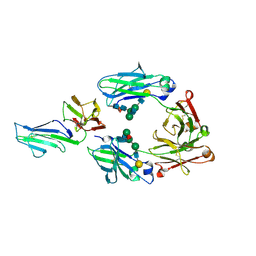 | | Crystal structure of IIb selective Fc variant, Fc(P238D), in complex with FcgRIIb | | Descriptor: | Ig gamma-1 chain C region, Low affinity immunoglobulin gamma Fc region receptor II-b, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kadono, S, Mimoto, F, Katada, H, Igawa, T, Kuramochi, T, Muraoka, M, Wada, Y, Haraya, K, Miyazaki, T, Hattori, K. | | Deposit date: | 2013-10-10 | | Release date: | 2013-11-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineered antibody Fc variant with selectively enhanced Fc gamma RIIb binding over both Fc gamma RIIaR131 and Fc gamma RIIaH131.
Protein Eng.Des.Sel., 26, 2013
|
|
8HHV
 
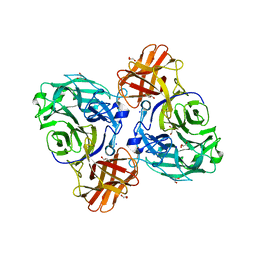 | | endo-alpha-D-arabinanase EndoMA1 from Microbacterium arabinogalactanolyticum | | Descriptor: | CALCIUM ION, GLYCEROL, SODIUM ION, ... | | Authors: | Nakashima, C, Li, J, Arakawa, T, Yamada, C, Ishiwata, A, Fujita, K, Fushinobu, S. | | Deposit date: | 2022-11-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and characterization of endo-alpha-, exo-alpha-, and exo-beta-D-arabinofuranosidases degrading lipoarabinomannan and arabinogalactan of mycobacteria.
Nat Commun, 14, 2023
|
|
