3OE5
 
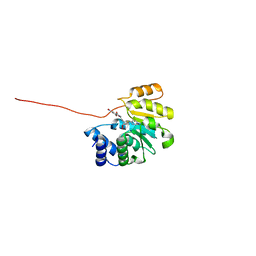 | | Rat catechol O-methyltransferase in complex with a catechol-type, pyridylsulfanyl-containing inhibitor - humanized form | | Descriptor: | Catechol O-methyltransferase, MAGNESIUM ION, N-[(E)-3-[(2R,3S,4R,5S)-3,4-dihydroxy-5-pyridin-4-ylsulfanyl-oxolan-2-yl]prop-2-enyl]-2,3-dihydroxy-5-nitro-benzamide | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2010-08-12 | | Release date: | 2011-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Molecular Recognition at the Active Site of Catechol-O-methyltransferase (COMT): Adenine Replacements in Bisubstrate Inhibitors
Chemistry, 17, 2011
|
|
8JBE
 
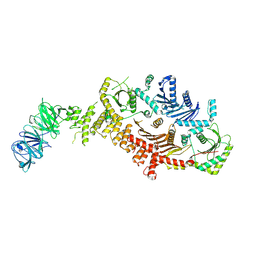 | | CryoEM Structure of metazoan Mon1-Ccz1-RMC1 complex | | Descriptor: | Caffeine, calcium, zinc sensitivity 1, ... | | Authors: | Jia, G.W, Yong, X, Su, Z.M, Jia, D. | | Deposit date: | 2023-05-08 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | CryoEM Structure of metazoan Mon1-Ccz1-RMC1 complex
To Be Published
|
|
2J6P
 
 | | STRUCTURE OF AS-SB REDUCTASE FROM LEISHMANIA MAJOR | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SB(V)-AS(V) REDUCTASE, ... | | Authors: | Bisacchi, D, Zhou, Y, Rosen, B.P, Mukhopadhyay, R, Bordo, D. | | Deposit date: | 2006-10-02 | | Release date: | 2007-10-02 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural characterization of the As/Sb reductase LmACR2 from Leishmania major.
J. Mol. Biol., 386, 2009
|
|
8J7P
 
 | |
5K55
 
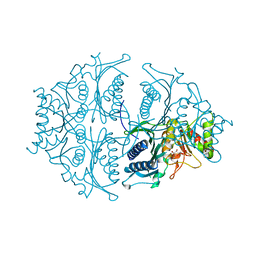 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state in complex with fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
5JYN
 
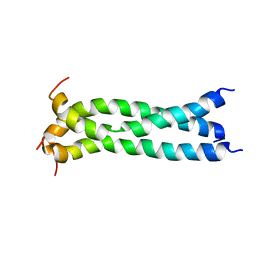 | | Structure of the transmembrane domain of HIV-1 gp41 in bicelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Dev, J, Fu, Q, Park, D, Chen, B, Chou, J.J. | | Deposit date: | 2016-05-14 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for membrane anchoring of HIV-1 envelope spike.
Science, 353, 2016
|
|
8J8G
 
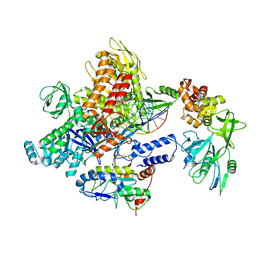 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 in complex with a DNA duplex and cidofovir diphosphate | | Descriptor: | CALCIUM ION, DNA (5'-D(P*AP*GP*CP*TP*GP*CP*TP*AP*TP*GP*AP*GP*AP*TP*TP*AP*AP*GP*TP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*GP*AP*TP*AP*AP*CP*TP*TP*AP*AP*TP*CP*TP*CP*AP*CP*AP*TP*AP*GP*CP*AP*GP*CP*T)-3'), ... | | Authors: | Xu, Y, Wu, Y, Wu, X, Zhang, Y, Yang, Y, Li, D, Yang, B, Gao, K, Zhang, Z, Dong, C. | | Deposit date: | 2023-05-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural basis of human mpox viral DNA replication inhibition by brincidofovir and cidofovir.
Int.J.Biol.Macromol., 270, 2024
|
|
8J8F
 
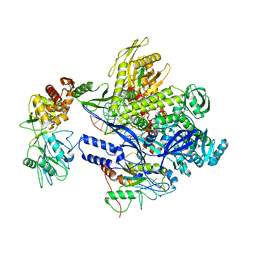 | | Monkeypox virus DNA replication holoenzyme F8, A22 and E4 in complex with a DNA duplex and dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*AP*GP*CP*TP*GP*CP*TP*AP*TP*GP*AP*GP*AP*TP*TP*AP*AP*GP*TP*TP*AP*T)-3'), ... | | Authors: | Xu, Y, Wu, Y, Wu, X, Zhang, Y, Yang, Y, Li, D, Yang, B, Gao, K, Zhang, Z, Dong, C. | | Deposit date: | 2023-05-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of human mpox viral DNA replication inhibition by brincidofovir and cidofovir.
Int.J.Biol.Macromol., 270, 2024
|
|
3DS6
 
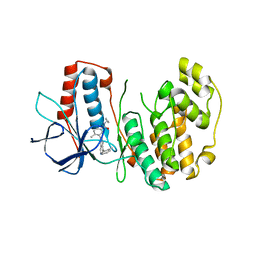 | | P38 complex with a phthalazine inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N-cyclopropyl-4-methyl-3-[1-(2-methylphenyl)phthalazin-6-yl]benzamide | | Authors: | Herberich, B, Syed, R, Li, V, Grosfeld, D. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of highly selective and potent p38 inhibitors based on a phthalazine scaffold.
J.Med.Chem., 51, 2008
|
|
5H46
 
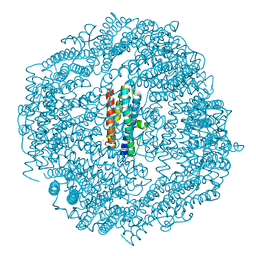 | | Mycobacterium smegmatis Dps1 mutant - F47E | | Descriptor: | DNA protection during starvation protein, FE (II) ION | | Authors: | Williams, S.M, Chandran, A.V, Vijayan, M, Chatterji, D. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Mutation Directs the Structural Switch of DNA Binding Proteins under Starvation to a Ferritin-like Protein Cage.
Structure, 25, 2017
|
|
3DMK
 
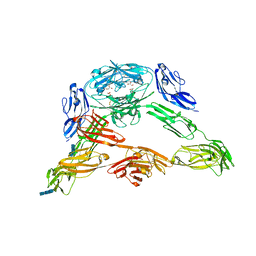 | | Crystal structure of Down Syndrome Cell Adhesion Molecule (DSCAM) isoform 1.30.30, N-terminal eight Ig domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule (DSCAM) isoform 1.30.30, ... | | Authors: | Sawaya, M.R, Wojtowicz, W.M, Eisenberg, D, Zipursky, S.L. | | Deposit date: | 2008-07-01 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.19 Å) | | Cite: | A double S shape provides the structural basis for the extraordinary binding specificity of Dscam isoforms.
Cell(Cambridge,Mass.), 134, 2008
|
|
7TLJ
 
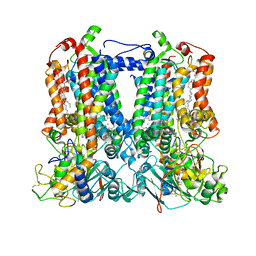 | | Rhodobacter sphaeroides Mitochondrial respiratory chain complex | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, (5S)-3-anilino-5-methyl-5-(6-phenoxypyridin-3-yl)-1,3-oxazolidine-2,4-dione, 14 kDa peptide of ubiquinol-cytochrome c2 oxidoreductase complex, ... | | Authors: | Xia, D, Zhou, F, Esser, L, Huang, R. | | Deposit date: | 2022-01-18 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Conformation Switch of Rieske ISP subunit is revealed by the Crystal Structure of Bacterial Cytochrome bc1 in Complex with Azoxystrobin
to be published
|
|
5FMZ
 
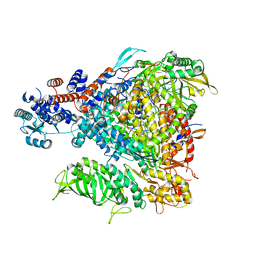 | | Crystal structure of Influenza B polymerase with bound 5' vRNA | | Descriptor: | 5'-R(*AP*GP*UP*AP*GP*UP*AP*AP*CP*AP*AP*GP)-3', POLYMERASE ACIDIC PROTEIN, POLYMERASE BASIC PROTEIN 2, ... | | Authors: | Guilligay, D, Cusack, S. | | Deposit date: | 2015-11-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of Pb2 Domains.
Mol.Cell, 61, 2016
|
|
2HGA
 
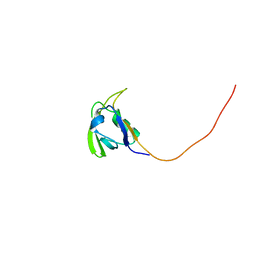 | | Solution NMR Structure of Conserved protein MTH1368, Northeast Structural Genomics Consortium Target TT821A | | Descriptor: | Conserved protein MTH1368 | | Authors: | Liu, G, Lin, Y, Parish, D, Shen, Y, Sukumaran, D, Yee, A, Semesi, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-26 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Conserved protein MTH1368, Northeast Structural Genomics Consortium Target TT821A
TO BE PUBLISHED
|
|
5FMQ
 
 | | Crystal structure of the mid, cap-binding, mid-link and 627 domains from avian influenza A virus polymerase PB2 subunit bound to M7GTP H32 crystal form | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, INFLUENZA A PB2 SUBUNIT | | Authors: | Thierry, E, Pflug, A, Hart, D, Cusack, S. | | Deposit date: | 2015-11-07 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of Pb2 Domains.
Mol.Cell, 61, 2016
|
|
8J47
 
 | | CryoEM Structure of 40-Residue Arctic (E22G) Beta-Amyloid Fibril Derived by Co-Analysis with Solid-State NMR | E22G Abeta40 | | Descriptor: | E22G Amyloid-beta | | Authors: | Tehrani, M.J, Matsuda, I, Yamagata, A, Matsunaga, T, Sato, M, Toyooka, K, Shirouzu, M, Ishii, Y, Kodama, Y, McElheny, D, Kobayashi, N. | | Deposit date: | 2023-04-19 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | E22G A beta 40 fibril structure and kinetics illuminate how A beta 40 rather than A beta 42 triggers familial Alzheimer's.
Nat Commun, 15, 2024
|
|
2KWI
 
 | | RalB-RLIP76 (RalBP1) complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RalA-binding protein 1, ... | | Authors: | Fenwick, R.B, Campbell, L.J, Rajasekar, K, Prasannan, S, Nietlispach, D, Camonis, J, Owen, D, Mott, H.R. | | Deposit date: | 2010-04-12 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The RalB-RLIP76 complex reveals a novel mode of ral-effector interaction
Structure, 18, 2010
|
|
3OZT
 
 | | Rat catechol O-methyltransferase in complex with a catechol-type, 4-oxo-pyridinyl-containing inhibitor - humanized form | | Descriptor: | Catechol O-methyltransferase, MAGNESIUM ION, N-[(E)-3-[(2R,3S,4R,5R)-3,4-dihydroxy-5-(4-oxopyridin-1-yl)oxolan-2-yl]prop-2-enyl]-2,3-dihydroxy-5-nitro-benzamide | | Authors: | Ehler, A, Schlatter, D, Stihle, M, Benz, J, Rudolph, M.G. | | Deposit date: | 2010-09-27 | | Release date: | 2011-03-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Molecular Recognition at the Active Site of Catechol-O-methyltransferase (COMT): Adenine Replacements in Bisubstrate Inhibitors
Chemistry, 17, 2011
|
|
5EI0
 
 | | Structure of RCL-cleaved vaspin (serpinA12) | | Descriptor: | Serpin A12 | | Authors: | Pippel, J, Kuettner, B.E, Ulbricht, D, Daberger, J, Schultz, S, Heiker, J.T, Strater, N. | | Deposit date: | 2015-10-29 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of cleaved vaspin (serpinA12).
Biol.Chem., 397, 2016
|
|
5K8Z
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 (pH 8.5) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Chlorite dismutase, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5K56
 
 | | Human muscle fructose-1,6-bisphosphatase in active R-state in complex with fructose-1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
8OEO
 
 | | Aspergillus niger ferulic acid decarboxylase (Fdc) V186C-A296C (DB4) variant in complex with prenylated flavin | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Roberts, G.W, Leys, D. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Aspergillus niger ferulic acid decarboxylase (Fdc) V186C-A296C (DB4) variant in complex with prenylated flavin
To Be Published
|
|
8OED
 
 | |
8OEH
 
 | | Aspergillus niger ferulic acid decarboxylase (Fdc) C122-S261C (DB3) variant in complex with prenylated flavin | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Roberts, G.W, Leys, D. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Aspergillus niger ferulic acid decarboxylase (Fdc) C122-S261C (DB3) variant in complex with prenylated flavin
To Be Published
|
|
2KOX
 
 | | NMR residual dipolar couplings identify long range correlated motions in the backbone of the protein ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Fenwick, R.B, Richter, B, Lee, D, Walter, K.F.A, Milovanovic, D, Becker, S, Lakomek, N.A, Griesinger, C, Salvatella, X. | | Deposit date: | 2009-10-02 | | Release date: | 2011-06-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Weak Long-Range Correlated Motions in a Surface Patch of Ubiquitin Involved in Molecular Recognition
J.Am.Chem.Soc., 2011
|
|
