4DPG
 
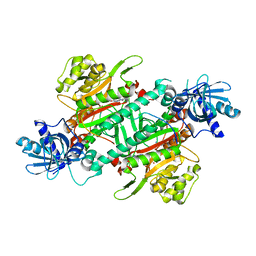 | | Crystal Structure of Human LysRS: P38/AIMP2 Complex I | | Descriptor: | ALANINE, Aminoacyl tRNA synthase complex-interacting multifunctional protein 2, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Fang, P, Wang, J, Bennett, S.P, Guo, M. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.844 Å) | | Cite: | Structural Switch of Lysyl-tRNA Synthetase between Translation and Transcription.
Mol.Cell, 49, 2013
|
|
2KKQ
 
 | | Solution NMR Structure of the Ig-like C2-type 2 Domain of Human Myotilin. Northeast Structural Genomics Target HR3158. | | Descriptor: | Myotilin | | Authors: | Rossi, P, Shastry, R, Ciccosanti, C, Hamilton, K, Xiao, R, Acton, T.B, Swapna, G.V.T, Nair, R, Everett, J.K, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Ig-like C2-type 2 Domain of Human Myotilin. Northeast Structural Genomics Target HR3158.
To be Published
|
|
8V4U
 
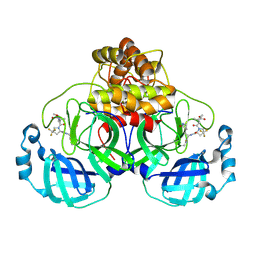 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | 3C-like proteinase nsp5, N-(methoxycarbonyl)-3-methyl-L-valyl-(4R)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-(trifluoromethyl)-L-prolinamide | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W. | | Deposit date: | 2023-11-29 | | Release date: | 2024-05-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | A Second-Generation Oral SARS-CoV-2 Main Protease Inhibitor Clinical Candidate for the Treatment of COVID-19.
J.Med.Chem., 67, 2024
|
|
4N7X
 
 | |
3OTQ
 
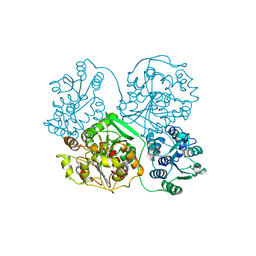 | | Soluble Epoxide Hydrolase in complex with pyrazole antagonist | | Descriptor: | Epoxide hydrolase 2, N-[4-(5-ethyl-3-pyridin-3-yl-1H-pyrazol-1-yl)phenyl]pyridine-3-carboxamide | | Authors: | Farrow, N.A. | | Deposit date: | 2010-09-13 | | Release date: | 2010-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Substituted pyrazoles as novel sEH antagonist: investigation of key binding interactions within the catalytic domain.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4N7W
 
 | | Crystal Structure of the sodium bile acid symporter from Yersinia frederiksenii | | Descriptor: | CITRIC ACID, Transporter, sodium/bile acid symporter family, ... | | Authors: | Zhou, X, Levin, E.J, Zhou, M, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2013-10-16 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural basis of the alternating-access mechanism in a bile acid transporter.
Nature, 505, 2013
|
|
2LNA
 
 | | Solution NMR Structure of the mitochondrial inner membrane domain (residues 164-251), FtsH_ext, from the paraplegin-like protein AFG3L2 from Homo sapiens, Northeast Structural Genomics Consortium Target HR6741A | | Descriptor: | AFG3-like protein 2 | | Authors: | Ramelot, T.A, Yang, Y, Lee, H, Janua, H, Kohan, E, Shastry, R, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and MD simulations of the AAA protease intermembrane space domain indicates peripheral membrane localization within the hexaoligomer.
Febs Lett., 587, 2013
|
|
2L0B
 
 | | Solution NMR structure of zinc finger domain of E3 ubiquitin-protein ligase praja-1 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) target HR4710B | | Descriptor: | E3 ubiquitin-protein ligase Praja-1, ZINC ION | | Authors: | Liu, G, Tong, S, Hamilton, K, Ciccosanti, C, Shastry, R, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of zinc finger domain of E3 ubiquitin-protein ligase protein praja-1 from Homo sapiens, northeast structural genomics consortium (NESG) target HR4710B
To be Published
|
|
6X8F
 
 | | Crystal structure of TYK2 with Compound 11 | | Descriptor: | Non-receptor tyrosine-protein kinase TYK2, [3-{4-[6-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrazin-4-yl]-1H-pyrazol-1-yl}-1-(2,2,2-trifluoroethyl)azetidin-3-yl]acetonitrile | | Authors: | Vajdos, F.F, Knafels, J.D. | | Deposit date: | 2020-06-01 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of Tyrosine Kinase 2 (TYK2) Inhibitor (PF-06826647) for the Treatment of Autoimmune Diseases.
J.Med.Chem., 63, 2020
|
|
8Q4E
 
 | |
8FUI
 
 | | HIV-1 wild type protease with GRL-02519A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
8FUJ
 
 | | HIV-1 wild type protease with GRL-03419A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group and 3,5-difluorophenylmethyl as the P1 group | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
6UVC
 
 | | Crystal structure of BCL-XL bound to compound 8: (R)-3-(Benzylthio)-2-(3-(2-((4'-chloro-[1,1'-biphenyl]-2-yl)methyl)-1,2,3,4-tetrahydroisoquinoline-6-carbonyl)-3-(4-methylbenzyl)ureido)propanoic acid | | Descriptor: | (R)-3-(Benzylthio)-2-(3-(2-((4'-chloro-[1,1'-biphenyl]-2-yl)methyl)-1,2,3,4-tetrahydroisoquinoline-6-carbonyl)-3-(4-methylbenzyl)ureido)propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1, ... | | Authors: | Roy, M.J, Birkinshaw, R, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
6UVD
 
 | | Crystal structure of BCL-XL bound to compound 2: (2R)-3-(Benzylsulfanyl)-2-({[(4-methylphenyl)methyl] [(4 phenylphenyl)carbonyl] carbamoyl}amino) propanoic acid | | Descriptor: | (2R)-3-(Benzylsulfanyl)-2-({[(4-methylphenyl)methyl] [(4 phenylphenyl)carbonyl] carbamoyl}amino) propanoic acid, 1,2-ETHANEDIOL, Bcl-2-like protein 1, ... | | Authors: | Roy, M.J, Birkinshaw, R, Lessene, G, Czabotar, P.E. | | Deposit date: | 2019-11-02 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Development of Potent Benzoylurea Inhibitors of BCL-X L and BCL-2.
J.Med.Chem., 64, 2021
|
|
8QK8
 
 | |
6Q7P
 
 | | Crystal structure of OE1.2 | | Descriptor: | 1,2-ETHANEDIOL, 1-PHENYLETHANONE, MAGNESIUM ION, ... | | Authors: | Levy, C.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Design and evolution of an enzyme with a non-canonical organocatalytic mechanism.
Nature, 570, 2019
|
|
7PPO
 
 | |
6Q7R
 
 | |
6Q7O
 
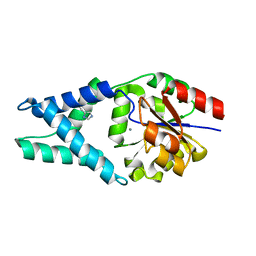 | | Crystal structure of OE1 | | Descriptor: | CALCIUM ION, OE1 | | Authors: | Levy, C.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and evolution of an enzyme with a non-canonical organocatalytic mechanism.
Nature, 570, 2019
|
|
6Q7N
 
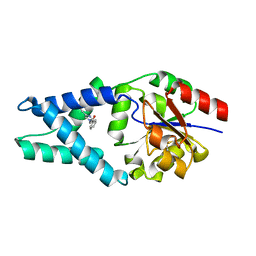 | |
6QIY
 
 | | CI-2, conformation 1 | | Descriptor: | Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Romero, A, Ruiz, F.M. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering protein assemblies with allosteric control via monomer fold-switching.
Nat Commun, 10, 2019
|
|
6Q7Q
 
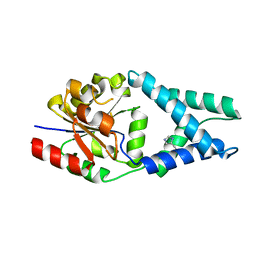 | | Crystal structure of OE1.3 | | Descriptor: | OE1.3 | | Authors: | Levy, C.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and evolution of an enzyme with a non-canonical organocatalytic mechanism.
Nature, 570, 2019
|
|
4MOT
 
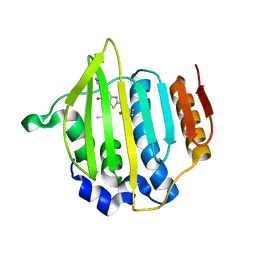 | | Structure of Streptococcus pneumonia pare in complex with AZ13072886 | | Descriptor: | 1-[4-(3-methylbutyl)-5-oxo-6-(pyridin-3-yl)-4,5-dihydro[1,3]thiazolo[5,4-b]pyridin-2-yl]-3-prop-2-en-1-ylurea, Topoisomerase IV subunit B | | Authors: | Ogg, D, Boriack-Sjodin, P.A. | | Deposit date: | 2013-09-12 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thiazolopyridone ureas as DNA gyrase B inhibitors: Optimization of antitubercular activity and efficacy.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6QIZ
 
 | | CI-2, conformation 2 | | Descriptor: | Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Romero, A, Ruiz, F.M. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Engineering protein assemblies with allosteric control via monomer fold-switching.
Nat Commun, 10, 2019
|
|
6XTT
 
 | |
