6Y39
 
 | |
3M63
 
 | |
3M62
 
 | |
3D6N
 
 | | Crystal Structure of Aquifex Dihydroorotase Activated by Aspartate Transcarbamoylase | | Descriptor: | Aspartate carbamoyltransferase, CITRATE ANION, Dihydroorotase, ... | | Authors: | Edwards, B.F.P. | | Deposit date: | 2008-05-20 | | Release date: | 2009-01-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dihydroorotase from the hyperthermophile Aquifiex aeolicus is activated by stoichiometric association with aspartate transcarbamoylase and forms a one-pot reactor for pyrimidine biosynthesis.
Biochemistry, 48, 2009
|
|
1OSY
 
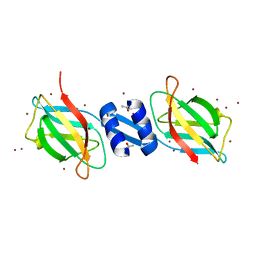 | | Crystal structure of FIP-Fve fungal immunomodulatory protein | | Descriptor: | BROMIDE ION, IMMUNOMODULATORY PROTEIN FIP-FVE | | Authors: | Palasingam, P, Joseph, J.S, Seow, S.V, Shai, V, Robinson, H, Chua, K.Y, Kolatkar, P.R. | | Deposit date: | 2003-03-20 | | Release date: | 2003-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A 1.7A structure of Fve, a member of the new fungal
immunomodulatory protein family
J.Mol.Biol., 332, 2003
|
|
4HXX
 
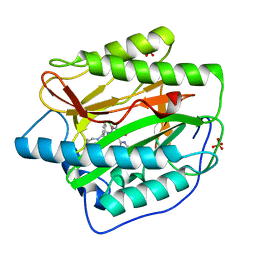 | | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1 | | Descriptor: | (1R)-N~2~-[5-chloro-2-(5-chloropyridin-2-yl)-6-methylpyrimidin-4-yl]-1-phenyl-N~1~-(4-phenylbutyl)ethane-1,2-diamine, COBALT (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Gabelli, S.B, Zhang, F, Liu, J, Amzel, L.M. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Pyridinylpyrimidines selectively inhibit human methionine aminopeptidase-1.
Bioorg.Med.Chem., 21, 2013
|
|
2EF7
 
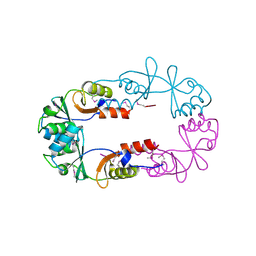 | | Crystal structure of ST2348, a hypothetical protein with CBS domains from Sulfolobus tokodaii strain7 | | Descriptor: | Hypothetical protein ST2348 | | Authors: | Agari, Y, Karthe, P, Kumarevel, T, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-21 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of ST2348, a CBS domain protein, from hyperthermophilic archaeon Sulfolobus tokodaii
Biochem.Biophys.Res.Commun., 375, 2008
|
|
3QQ7
 
 | |
3IPO
 
 | | Crystal structure of YnjE | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | Authors: | Haenzelmann, P, Kuper, J, Schindelin, H. | | Deposit date: | 2009-08-18 | | Release date: | 2009-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
3QIA
 
 | | Crystal structure of P-loop G237A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-26 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3IPP
 
 | | crystal structure of sulfur-free YnjE | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative thiosulfate sulfurtransferase ynjE, ... | | Authors: | Haenzelmann, P, Kuper, J, Schindelin, H. | | Deposit date: | 2009-08-18 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
3QG1
 
 | | Crystal structure of P-loop G239A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, V-type ATP synthase alpha chain | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Kumar, A, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-24 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3R3M
 
 | |
3QJY
 
 | | Crystal structure of P-loop G234A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-31 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3QQ8
 
 | |
1FFU
 
 | | CARBON MONOXIDE DEHYDROGENASE FROM HYDROGENOPHAGA PSEUDOFLAVA WHICH LACKS THE MO-PYRANOPTERIN MOIETY OF THE MOLYBDENUM COFACTOR | | Descriptor: | CUTL, MOLYBDOPROTEIN OF CARBON MONOXIDE DEHYDROGENASE, CUTM, ... | | Authors: | Haenzelmann, P, Dobbek, H, Gremer, L, Huber, R, Meyer, O. | | Deposit date: | 2000-07-26 | | Release date: | 2000-09-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase.
J.Mol.Biol., 301, 2000
|
|
1FFV
 
 | | CARBON MONOXIDE DEHYDROGENASE FROM HYDROGENOPHAGA PSEUDOFLAVA | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), CUTL, MOLYBDOPROTEIN OF CARBON MONOXIDE DEHYDROGENASE, ... | | Authors: | Haenzelmann, P, Dobbek, H, Gremer, L, Huber, R, Meyer, O. | | Deposit date: | 2000-07-26 | | Release date: | 2000-09-15 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase.
J.Mol.Biol., 301, 2000
|
|
5O36
 
 | | Japanese encephalitis virus non-structural protein 1' C-terminal domain | | Descriptor: | Japanese encephalitis virus non-structural protein 1' (NS1'),Japanese encephalitis virus non-structural protein 1' (NS1'), N-PROPANOL, SULFATE ION | | Authors: | Thanalai, P, Wright, G.S.A, Antonyuk, S.V. | | Deposit date: | 2017-05-23 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Study of the C-Terminal Domain of Nonstructural Protein 1 from Japanese Encephalitis Virus.
J. Virol., 92, 2018
|
|
4H0F
 
 | |
3TIW
 
 | | Crystal structure of p97N in complex with the C-terminus of gp78 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase AMFR, Transitional endoplasmic reticulum ATPase | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2011-08-22 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | The Structural and Functional Basis of the p97/Valosin-containing Protein (VCP)-interacting Motif (VIM): MUTUALLY EXCLUSIVE BINDING OF COFACTORS TO THE N-TERMINAL DOMAIN OF p97.
J.Biol.Chem., 286, 2011
|
|
1TV7
 
 | | Structure of the S-adenosylmethionine dependent Enzyme MoaA | | Descriptor: | IRON/SULFUR CLUSTER, Molybdenum cofactor biosynthesis protein A, SULFATE ION | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2004-06-28 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
6KRF
 
 | |
6KRC
 
 | | An X-ray structure of ferric F43Y/F46S sperm whale myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yuan, H, Lin, Y.W. | | Deposit date: | 2019-08-21 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A Catalytic Binding Site Together with a Distal Tyr in MyoglobinAffords Catalytic Efficiencies Similar to Natural Peroxidases.
Acs Catalysis, 10, 2020
|
|
7DRV
 
 | | Structural basis of SARS-CoV-2-closely-related bat coronavirus RaTG13 to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liu, K.F, Pan, X.Q, Li, L.J, Feng, Y, Meng, Y.M, Zhang, Y.F, Wu, L.L, Chen, Q, Zheng, A.Q, Song, C.L, Jia, Y.F, Niu, S, Qiao, C.P, Zhao, X, Ma, D.L, Ma, X.P, Tan, S.G, Qi, J.X, Gao, G.F, Wang, Q.H. | | Deposit date: | 2020-12-29 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species.
Cell, 184, 2021
|
|
7DDP
 
 | | Cryo-EM structure of human ACE2 and GX/P2V/2017 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Niu, S, Wang, J, Wang, H.W, Qi, J.X, Wang, Q.H, Gao, G.F. | | Deposit date: | 2020-10-29 | | Release date: | 2021-05-19 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin.
Embo J., 40, 2021
|
|
