7C34
 
 | |
3RIY
 
 | |
3TFK
 
 | | 42F3-p4B10/H2-Ld | | Descriptor: | 42F3 alpha, 42F3 beta, H2-Ld SBM2, ... | | Authors: | Adams, J.J, Kranz, D.M, Garcia, K.C. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
3TF7
 
 | | 42F3 QL9/H2-Ld complex | | Descriptor: | 42F3 Mut7 scFv (42F3 alpha chain, linker, 42F3 beta chain), ... | | Authors: | Adams, J.J, Kranz, D.M, Garcia, K.C. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
3RIG
 
 | |
3TJH
 
 | | 42F3-p3A1/H2-Ld complex | | Descriptor: | 42F3 alpha, 42F3 beta, H2-Ld SBM2, ... | | Authors: | Adams, J.J, Kruse, A, Kranz, D.M, Garcia, K.C. | | Deposit date: | 2011-08-24 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
3TPU
 
 | | 42F3 p5E8/H2-Ld complex | | Descriptor: | 1,2-ETHANEDIOL, 42F3 alpha, 42F3 beta, ... | | Authors: | Adams, J.J, Kranz, D.M, Garcia, K.C. | | Deposit date: | 2011-09-08 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
6A5M
 
 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with SAM, form 2 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6, S-ADENOSYLMETHIONINE, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A5N
 
 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with methylated DNA | | Descriptor: | DNA (5'-D(*CP*AP*CP*TP*GP*CP*TP*GP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*GP*AP*GP*TP*AP*CP*TP*(5CM)P*AP*GP*CP*AP*GP*T)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6A5K
 
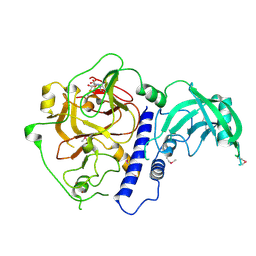 | | Crystal structure of Arabidopsis thaliana SUVH6 in complex with SAM, form 1 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6, S-ADENOSYLMETHIONINE, ... | | Authors: | Li, X, Du, J. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6AVF
 
 | | Crystal structure of the KFJ5 TCR-NY-ESO-1-HLA-B*07:02 complex | | Descriptor: | ALA-PRO-ARG-GLY-PRO-HIS-GLY-GLY-ALA-ALA-SER-GLY-LEU, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gully, B.S, Gras, S, Rossjohn, J. | | Deposit date: | 2017-09-02 | | Release date: | 2018-02-28 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2.028 Å) | | Cite: | Divergent T-cell receptor recognition modes of a HLA-I restricted extended tumour-associated peptide.
Nat Commun, 9, 2018
|
|
6AT6
 
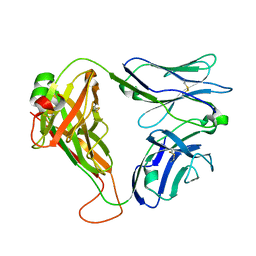 | | Crystal structure of the KFJ5 TCR | | Descriptor: | T-cell receptor alpha variable 4, T-cell receptor, sp3.4 alpha chain chimera, ... | | Authors: | Gully, B.S, Rossjohn, J. | | Deposit date: | 2017-08-28 | | Release date: | 2018-02-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.417 Å) | | Cite: | Divergent T-cell receptor recognition modes of a HLA-I restricted extended tumour-associated peptide.
Nat Commun, 9, 2018
|
|
6AVG
 
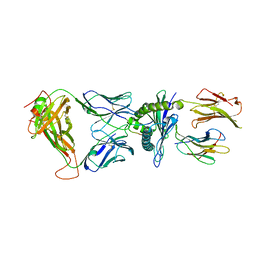 | | Crystal structure of the KFJ37 TCR-NY-ESO-1-HLA-B*07:02 complex | | Descriptor: | ALA-PRO-ARG-GLY-PRO-HIS-GLY-GLY-ALA-ALA-SER-GLY-LEU, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gully, B.S, Gras, S, Rossjohn, J. | | Deposit date: | 2017-09-02 | | Release date: | 2018-02-28 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Divergent T-cell receptor recognition modes of a HLA-I restricted extended tumour-associated peptide.
Nat Commun, 9, 2018
|
|
6AT5
 
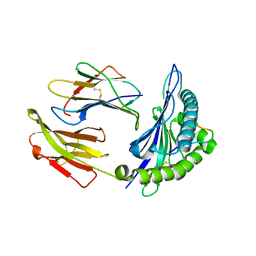 | |
7LGD
 
 | | HLA-B*07:02 in complex with SARS-CoV-2 nucleocapsid peptide N105-113 | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Szeto, C, Chatzileontiadou, D.S.M. | | Deposit date: | 2021-01-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | CD8 + T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses.
Immunity, 54, 2021
|
|
7LGT
 
 | | HLA-B*07:02 in complex with 229E-derived coronavirus nucleocapsid peptide N75-83 | | Descriptor: | BROMIDE ION, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Gras, S, Szeto, C, Chatzileontiadou, D.S.M. | | Deposit date: | 2021-01-21 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | CD8 + T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses.
Immunity, 54, 2021
|
|
7C2L
 
 | | S protein of SARS-CoV-2 in complex bound with 4A8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Guo, Y.Y, Li, Y.N, Xia, L, Zhou, Q. | | Deposit date: | 2020-05-08 | | Release date: | 2020-07-01 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2.
Science, 369, 2020
|
|
6LXH
 
 | | Staphylococcus aureus surface protein-sdrc | | Descriptor: | Ser-Asp rich fibrinogen-binding, bone sialoprotein-binding protein | | Authors: | Pi, Y, Ji, Q. | | Deposit date: | 2020-02-11 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis ofStaphylococcus aureusSurface Protein SdrC.
Biochemistry, 59, 2020
|
|
6LXS
 
 | | CWA protein-SdrC | | Descriptor: | CALCIUM ION, GLYCEROL, Ser-Asp rich fibrinogen-binding, ... | | Authors: | Pi, Y, Ji, Q. | | Deposit date: | 2020-02-11 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Basis ofStaphylococcus aureusSurface Protein SdrC.
Biochemistry, 59, 2020
|
|
8HLT
 
 | | The co-crystal structure of DYRK2 with YK-2-99B | | Descriptor: | (6-{[(4P)-4-(1,3-benzothiazol-5-yl)-5-fluoropyrimidin-2-yl]amino}pyridin-3-yl)(piperazin-1-yl)methanone, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Shen, H.T, Xiao, Y.B, Yuan, K, Yang, P, Li, Q.N. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Potent DYRK2 Inhibitors with High Selectivity, Great Solubility, and Excellent Safety Properties for the Treatment of Prostate Cancer.
J.Med.Chem., 66, 2023
|
|
6MT3
 
 | |
6MTL
 
 | |
6MT5
 
 | |
6MT4
 
 | |
6MTM
 
 | | Crystal Structure of EM2 TCR in complex with HLA-B*37:01-NP338 | | Descriptor: | Beta-2-microglobulin, EM2 TCR alpha chain, EM2 TCR beta chain, ... | | Authors: | Gras, S. | | Deposit date: | 2018-10-19 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Broad CD8+T cell cross-recognition of distinct influenza A strains in humans.
Nat Commun, 9, 2018
|
|
