1NH6
 
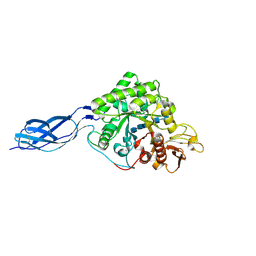 | | Structure of S. marcescens chitinase A, E315L, complex with hexasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, chitinase A | | Authors: | Aronson Jr, N.N, Halloran, B.A, Alexyev, M.F, Amable, L, Madura, J.D, Pasupulati, L, Worth, C, Van Roey, P. | | Deposit date: | 2002-12-18 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Family 18 chitinase-oligosaccharide substrate interaction: subsite preference and anomer selectivity of Serratia marcescens chitinase A.
Biochem.J., 376, 2003
|
|
1L9X
 
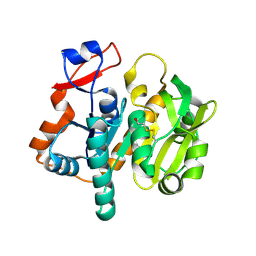 | | Structure of gamma-Glutamyl Hydrolase | | Descriptor: | BETA-MERCAPTOETHANOL, gamma-glutamyl hydrolase | | Authors: | Li, H, Ryan, T.J, Chave, K.J, Van Roey, P. | | Deposit date: | 2002-03-26 | | Release date: | 2002-04-10 | | Last modified: | 2021-04-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three-dimensional structure of human gamma -glutamyl hydrolase. A class I glatamine amidotransferase adapted for a complex substate.
J.Biol.Chem., 277, 2002
|
|
1C3F
 
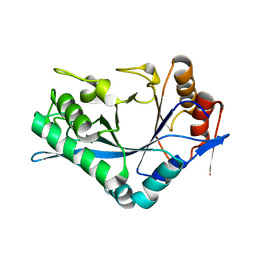 | | Endo-Beta-N-Acetylglucosaminidase H, D130N Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-27 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1C90
 
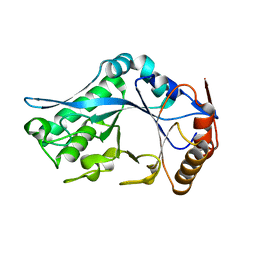 | | Endo-Beta-N-Acetylglucosaminidase H, E132Q Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H | | Authors: | Rao, V, Tao, C, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Assp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1C92
 
 | | Endo-Beta-N-Acetylglucosaminidase H, E132A Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1C8X
 
 | | Endo-Beta-N-Acetylglucosaminidase H, D130E Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H, PHOSPHATE ION | | Authors: | Rao, V, Tao, C, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1C91
 
 | | Endo-Beta-N-Acetylglucosaminidase H, E132D | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1C8Y
 
 | | Endo-Beta-N-Acetylglucosaminidase H, D130A Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H, ZINC ION | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1C93
 
 | | Endo-Beta-N-Acetylglucosaminidase H, D130N/E132Q Double Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-30 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
1VQ2
 
 | | CRYSTAL STRUCTURE OF T4-BACTERIOPHAGE DEOXYCYTIDYLATE DEAMINASE, MUTANT R115E | | Descriptor: | 3,4-DIHYDRO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, DEOXYCYTIDYLATE DEAMINASE, ZINC ION | | Authors: | Almog, R, Maley, F, Maley, G.F, Maccoll, R, Van Roey, P. | | Deposit date: | 2004-12-15 | | Release date: | 2004-12-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-Dimensional Structure of the R115E Mutant of T4-Bacteriophage 2'-Deoxycytidylate Deaminase
Biochemistry, 43, 2004
|
|
1YW5
 
 | | Peptidyl-prolyl isomerase ESS1 from Candida albicans | | Descriptor: | peptidyl prolyl cis/trans isomerase | | Authors: | Li, Z, Li, H, Devasahayam, G, Gemmill, T, Chaturvedi, V, Hanes, S.D, Van Roey, P. | | Deposit date: | 2005-02-17 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of the Candida albicans Ess1 Prolyl Isomerase Reveals a Well-Ordered Linker that Restricts Domain Mobility
Biochemistry, 44, 2005
|
|
4GIG
 
 | |
1B02
 
 | | CRYSTAL STRUCTURE OF THYMIDYLATE SYNTHASE A FROM BACILLUS SUBTILIS | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, PROTEIN (THYMIDYLATE SYNTHASE) | | Authors: | Fox, K.M, Maley, F, Garibian, A, Changchien, L, Vanroey, P. | | Deposit date: | 1998-11-16 | | Release date: | 1999-03-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of thymidylate synthase A from Bacillus subtilis.
Protein Sci., 8, 1999
|
|
4LW5
 
 | | Crystal structure of all-trans green fluorescent protein | | Descriptor: | Green fluorescent protein | | Authors: | Rosenman, D.J, Huang, Y.-M, Xia, K, Vanroey, P, Colon, W, Bystroff, C. | | Deposit date: | 2013-07-26 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Green-lighting green fluorescent protein: Faster and more efficient folding by eliminating a cis-trans peptide isomerization event.
Protein Sci., 23, 2014
|
|
2ZAL
 
 | | Crystal structure of E. coli isoaspartyl aminopeptidase/L-asparaginase in complex with L-aspartate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ASPARTIC ACID, CALCIUM ION, ... | | Authors: | Michalska, K, Brzezinski, K, Jaskolski, M. | | Deposit date: | 2007-10-07 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of isoaspartyl aminopeptidase in complex with L-aspartate
J.Biol.Chem., 280, 2005
|
|
1PGS
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF PNGASE F, A GLYCOSYLASPARAGINASE FROM FLAVOBACTERIUM MENINGOSEPTICUM | | Descriptor: | PEPTIDE-N(4)-(N-ACETYL-BETA-D-GLUCOSAMINYL)ASPARAGINE AMIDASE F | | Authors: | Norris, G.E, Stillman, T.J, Anderson, B.F, Baker, E.N. | | Deposit date: | 1994-10-06 | | Release date: | 1995-01-26 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of PNGase F, a glycosylasparaginase from Flavobacterium meningosepticum.
Structure, 2, 1994
|
|
1JN9
 
 | |
1K2X
 
 | |
1ZN1
 
 | | Coordinates of RRF fitted into Cryo-EM map of the 70S post-termination complex | | Descriptor: | 30S ribosomal protein S12, Ribosome recycling factor, ribosomal 16S RNA, ... | | Authors: | Gao, N, Zavialov, A.V, Li, W, Sengupta, J, Valle, M, Gursky, R.P, Ehrenberg, M, Frank, J. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies.
Mol.Cell, 18, 2005
|
|
