7NIN
 
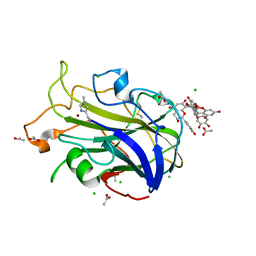 | | X-ray crystal structure of LsAA9A - CinnamtanninB1 soak | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Auxiliary activity 9, ... | | Authors: | Frandsen, K.E.H, Tokin, R, Skov, L, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-02-12 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of lytic polysaccharide monooxygenase by natural plant extracts.
New Phytol., 232, 2021
|
|
7NIM
 
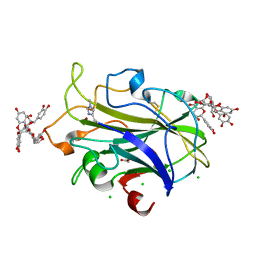 | | X-ray crystal structure of LsAA9A - cinnamon extract soak | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Auxiliary activity 9, ... | | Authors: | Frandsen, K.E.H, Tokin, R, Skov, L, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-02-12 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Inhibition of lytic polysaccharide monooxygenase by natural plant extracts.
New Phytol., 232, 2021
|
|
2AB6
 
 | |
5U3I
 
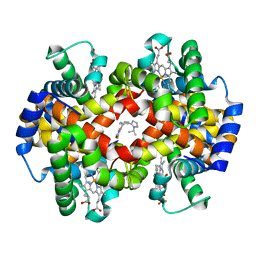 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN S (LIGANDED SICKLE CELL HEMOGLOBIN) COMPLEXED WITH GBT compound 31 | | Descriptor: | 2-methoxy-5-({2-[1-(propan-2-yl)-1H-pyrazol-5-yl]pyridin-3-yl}methoxy)pyridine-4-carbaldehyde, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Partridge, J.R, Choy, R.M, Li, Z, Metcalf, B. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of GBT440, an Orally Bioavailable R-State Stabilizer of Sickle Cell Hemoglobin.
ACS Med Chem Lett, 8, 2017
|
|
5UFJ
 
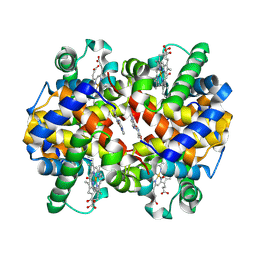 | | Crystal Structure of Carbonmonoxy Hemoglobin S (Liganded Sickle Cell Hemoglobin) Complexed with GBT Compound 6 | | Descriptor: | 5-[(imidazo[1,2-a]pyridin-8-yl)methoxy]-2-methoxypyridine-4-carbaldehyde, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Partridge, J.R, Choy, R.M, Li, Z, Metcalf, B. | | Deposit date: | 2017-01-04 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of GBT440, an Orally Bioavailable R-State Stabilizer of Sickle Cell Hemoglobin.
ACS Med Chem Lett, 8, 2017
|
|
5TGL
 
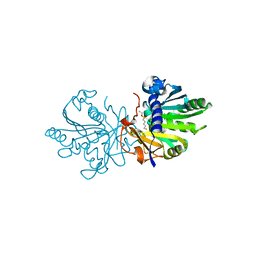 | | A MODEL FOR INTERFACIAL ACTIVATION IN LIPASES FROM THE STRUCTURE OF A FUNGAL LIPASE-INHIBITOR COMPLEX | | Descriptor: | LIPASE, N-HEXYLPHOSPHONATE ETHYL ESTER | | Authors: | Brzozowski, A.M, Derewenda, U, Derewenda, Z.S, Dodson, G.G, Lawson, D, Turkenburg, J.P, Bjorkling, F, Huge-Jensen, B, Patkar, S.R, Thim, L. | | Deposit date: | 1991-10-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex.
Nature, 351, 1991
|
|
4TGL
 
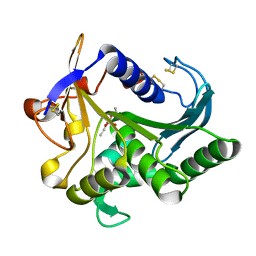 | | CATALYSIS AT THE INTERFACE: THE ANATOMY OF A CONFORMATIONAL CHANGE IN A TRIGLYCERIDE LIPASE | | Descriptor: | DIETHYL PHOSPHONATE, TRIACYL-GLYCEROL ACYLHYDROLASE | | Authors: | Derewenda, U, Brzozowski, A.M, Lawson, D, Derewenda, Z.S. | | Deposit date: | 1991-07-29 | | Release date: | 1993-07-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalysis at the interface: the anatomy of a conformational change in a triglyceride lipase.
Biochemistry, 31, 1992
|
|
5MZ8
 
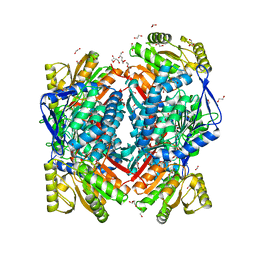 | | Crystal structure of aldehyde dehydrogenase 21 (ALDH21) from Physcomitrella patens in complex with the reaction product succinate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kopecny, D, Vigouroux, A, Briozzo, P, Morera, S. | | Deposit date: | 2017-01-31 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The ALDH21 gene found in lower plants and some vascular plants codes for a NADP(+) -dependent succinic semialdehyde dehydrogenase.
Plant J., 92, 2017
|
|
5MZ5
 
 | | Crystal structure of aldehyde dehydrogenase 21 (ALDH21) from Physcomitrella patens in its apoform | | Descriptor: | 1,2-ETHANEDIOL, ALDH21), DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kopecny, D, Koncitikova, R, Briozzo, P, Morera, S. | | Deposit date: | 2017-01-30 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The ALDH21 gene found in lower plants and some vascular plants codes for a NADP(+) -dependent succinic semialdehyde dehydrogenase.
Plant J., 92, 2017
|
|
5N5S
 
 | | Crystal structure of aldehyde dehydrogenase 21 (ALDH21) from Physcomitrella patens in complex with NADP+ | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase 21 (ALDH21), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kopecny, D, Vigouroux, A, Briozzo, P, Morera, S. | | Deposit date: | 2017-02-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The ALDH21 gene found in lower plants and some vascular plants codes for a NADP(+) -dependent succinic semialdehyde dehydrogenase.
Plant J., 92, 2017
|
|
2NQ3
 
 | | Crystal structure of the C2 Domain of Human Itchy Homolog E3 Ubiquitin Protein Ligase | | Descriptor: | CHLORIDE ION, Itchy homolog E3 ubiquitin protein ligase | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-30 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The C2 Domain of Human Itchy Homolog E3 Ubiquitin Protein Ligase
To be Published
|
|
2NSQ
 
 | | Crystal structure of the C2 domain of the human E3 ubiquitin-protein ligase NEDD4-like protein | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase NEDD4-like protein, GLYCEROL | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-06 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The C2 domain of the human E3 ubiquitin-protein ligase NEDD4-like protein
To be Published
|
|
