7D81
 
 | |
7D7Y
 
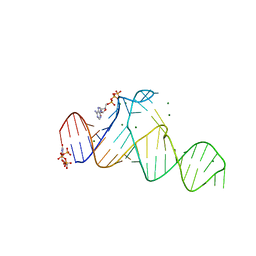 | |
7ELR
 
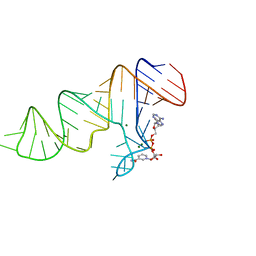
 | | Crystal structure of xanthine riboswitch with xanthine | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NMT1 (46-MER), ... | | Authors: | Xu, X.C, Ren, A.M. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Insights into xanthine riboswitch structure and metal ion-mediated ligand recognition.
Nucleic Acids Res., 49, 2021
|
|
7ELS
 
 | | Crystal structure of xanthine riboswitch with 8-azaxanthine | | Descriptor: | 8-AZAXANTHINE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xu, X.C, Ren, A.M. | | Deposit date: | 2021-04-12 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into xanthine riboswitch structure and metal ion-mediated ligand recognition.
Nucleic Acids Res., 49, 2021
|
|
7ELQ
 
 | |
7ELP
 
 | |
7D7V
 
 | |
7D7Z
 
 | |
7EOK
 
 | | Crystal structure of the Pepper aptamer in complex with HBC485 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[5-[2-(dimethylamino)ethyl-methyl-amino]pyrazin-2-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOO
 
 | | Crystal structure of the Pepper aptamer in complex with HBC525 | | Descriptor: | (~{E})-2-(1,3-benzoxazol-2-yl)-3-[4-[2-hydroxyethyl(methyl)amino]phenyl]prop-2-enenitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOP
 
 | | Crystal structure of the Pepper aptamer in complex with HBC620 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]thieno[3,2-b]thiophen-2-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOL
 
 | | Crystal structure of the Pepper aptamer in complex with HBC497 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]pyrazin-2-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOJ
 
 | |
7EOI
 
 | |
7EOG
 
 | |
7EON
 
 | | Crystal structure of the Pepper aptamer in complex with HBC514 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[4-[2-(dimethylamino)ethyl-methyl-amino]phenyl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOM
 
 | | Crystal structure of the Pepper aptamer in complex with HBC508 | | Descriptor: | 4-[(~{Z})-1-cyano-2-[6-[2-hydroxyethyl(methyl)amino]pyridin-3-yl]ethenyl]benzenecarbonitrile, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
7EOH
 
 | | Crystal structure of the Pepper aptamer in complex with HBC | | Descriptor: | 4-[(~{Z})-1-cyano-2-[4-[2-hydroxyethyl(methyl)amino]phenyl]ethenyl]benzenecarbonitrile, MAGNESIUM ION, Pepper (49-MER) | | Authors: | Huang, K.Y, Ren, A.M. | | Deposit date: | 2021-04-22 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.637 Å) | | Cite: | Structure-based investigation of fluorogenic Pepper aptamer.
Nat.Chem.Biol., 17, 2021
|
|
8CZ9
 
 | |
4YIS
 
 | | Crystal Structure of LAGLIDADG Meganuclease I-CpaMI Bound to Uncleaved DNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (28-MER), ... | | Authors: | Hallinan, J.P, Kaiser, B.K, Stoddard, B.L. | | Deposit date: | 2015-03-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Indirect DNA Sequence Recognition and Its Impact on Nuclease Cleavage Activity.
Structure, 24, 2016
|
|
4YIT
 
 | |
4YHX
 
 | |
4AA9
 
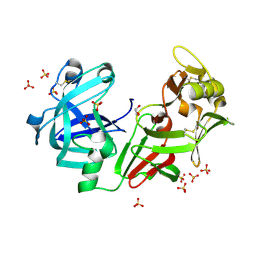 | | Camel chymosin at 1.6A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHYMOSIN, GLYCEROL, ... | | Authors: | Langholm Jensen, J, Molgaard, A, Navarro Poulsen, J.C, van den Brink, J.M, Harboe, M, Simonsen, J.B, Qvist, K.B, Larsen, S. | | Deposit date: | 2011-11-30 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Camel and Bovine Chymosin: The Relationship between Their Structures and Cheese-Making Properties.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4Z1Z
 
 | |
4Z20
 
 | |
