7VS9
 
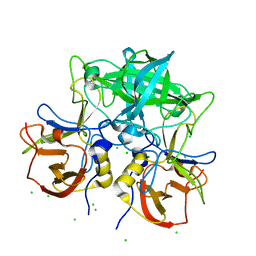 | | Crystal structure of P domain from norovirus GI.9 capsid protein in complex with Lewis x antigen. | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, VP1, ... | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2021-10-26 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Lewis fucose is a key moiety for the recognition of histo-blood group antigens by GI.9 norovirus, as revealed by structural analysis.
Febs Open Bio, 12, 2022
|
|
7VS8
 
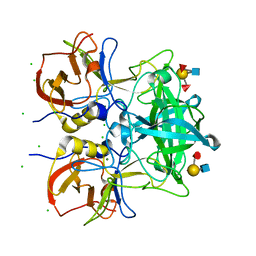 | | Crystal structure of P domain from norovirus GI.9 capsid protein in complex with Lewis b antigen. | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, VP1, ... | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2021-10-26 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lewis fucose is a key moiety for the recognition of histo-blood group antigens by GI.9 norovirus, as revealed by structural analysis.
Febs Open Bio, 12, 2022
|
|
7CD1
 
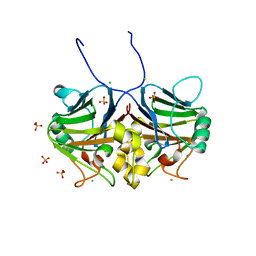 | | Crystal structure of inhibitory Smad, Smad7 | | Descriptor: | CHLORIDE ION, Mothers against decapentaplegic homolog 7, SULFATE ION | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for inhibitory effects of Smad7 on TGF-beta family signaling.
J.Struct.Biol., 212, 2020
|
|
1WEK
 
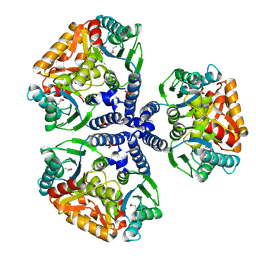 | | Crystal structure of the conserved hypothetical protein TT1465 from Thermus thermophilus HB8 | | Descriptor: | PHOSPHATE ION, hypothetical protein TT1465 | | Authors: | Kukimoto-Niino, M, Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-25 | | Release date: | 2004-11-25 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of possible lysine decarboxylases from Thermus thermophilus HB8
Protein Sci., 13, 2004
|
|
1WDV
 
 | | Crystal structure of hypothetical protein APE2540 | | Descriptor: | hypothetical protein APE2540 | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-18 | | Release date: | 2004-11-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a putative trans-editing enzyme for prolyl-tRNA synthetase from Aeropyrum pernix K1 at 1.7 A resolution.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
7EJL
 
 | |
7EJN
 
 | |
7EJM
 
 | |
1UF3
 
 | | Crystal structure of TT1561 of thermus thermophilus HB8 | | Descriptor: | CALCIUM ION, hypothetical protein TT1561 | | Authors: | Kato-Murayama, M, Shirouzu, M, Terada, T, Murayama, K, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-23 | | Release date: | 2003-11-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of TT1561 of thermus thermophilus HB8
To be Published
|
|
5Y7Z
 
 | | Complex structure of cyclin G-associated kinase with gefitinib | | Descriptor: | Cyclin-G-associated kinase, Gefitinib, NANOBODY, ... | | Authors: | Ohbayashi, N, Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2017-08-18 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structural Basis for the Inhibition of Cyclin G-Associated Kinase by Gefitinib.
ChemistryOpen, 7, 2018
|
|
5Y80
 
 | | Complex structure of cyclin G-associated kinase with gefitinib | | Descriptor: | Cyclin-G-associated kinase, Gefitinib, NANOBODY | | Authors: | Ohbayashi, N, Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2017-08-18 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Inhibition of Cyclin G-Associated Kinase by Gefitinib.
ChemistryOpen, 7, 2018
|
|
1WD6
 
 | |
1WFX
 
 | | Crystal Structure of APE0204 from Aeropyrum pernix | | Descriptor: | CHLORIDE ION, Probable RNA 2'-phosphotransferase, ZINC ION | | Authors: | Kato-Murayama, M, Bessho, Y, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2004-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the RNA 2'-Phosphotransferase from Aeropyrum pernix K1
J.Mol.Biol., 348, 2005
|
|
5C2K
 
 | | Crystal structure of the fusion protein linked by RhoA and the GAP domain of MgcRacGAP | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Murayama, K, Kato-Murayama, M, Hosaka, T, Kitamura, T, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2015-06-16 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural basis of G-protein target alternation of MgcRacGAP by phospholylation
To Be Published
|
|
5C2J
 
 | | Complex structure of the GAP domain of MgcRacGAP and Cdc42 | | Descriptor: | ALUMINUM FLUORIDE, Cell division control protein 42 homolog, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Murayama, K, Kato-Murayama, M, Hosaka, T, Kitamura, T, Yokoyama, S, Shirouzu, M. | | Deposit date: | 2015-06-16 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of G-protein target alternation of MgcRacGAP by phospholylation
To Be Published
|
|
4YN3
 
 | | Crystal structure of Cucumisin complex with pro-peptide | | Descriptor: | CHLORIDE ION, Cucumisin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Murayama, K, Kato-Murayama, M, Yokoyama, S, Arima, K, Shirouzu, M. | | Deposit date: | 2015-03-09 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of cucumisin protease activity regulation by its propeptide
J. Biochem., 161, 2017
|
|
2Z0O
 
 | | Crystal structure of APPL1-BAR-PH domain | | Descriptor: | DCC-interacting protein 13-alpha | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of APPL1-BAR-PH domain
To be Published
|
|
2Z0V
 
 | | Crystal structure of BAR domain of Endophilin-III | | Descriptor: | SH3-containing GRB2-like protein 3 | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of BAR domain of Endophilin-III
To be Published
|
|
2Z0Z
 
 | | Crystal structure of putative acetyltransferase | | Descriptor: | Putative uncharacterized protein TTHA1799, SULFATE ION | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Genetic Encoding of 3-Iodo-l-Tyrosine in Escherichia coli for Single-Wavelength Anomalous Dispersion Phasing in Protein Crystallography
Structure, 17, 2009
|
|
2Z0T
 
 | | Crystal structure of hypothetical protein PH0355 | | Descriptor: | Putative uncharacterized protein PH0355 | | Authors: | Murayama, K, Takemoto, C, Kato-Murayama, M, Kawazoe, M, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hypothetical protein PH0355
To be Published
|
|
2Z10
 
 | | Crystal structure of putative acetyltransferase | | Descriptor: | Ribosomal-protein-alanine acetyltransferase | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Genetic Encoding of 3-Iodo-l-Tyrosine in Escherichia coli for Single-Wavelength Anomalous Dispersion Phasing in Protein Crystallography
Structure, 17, 2009
|
|
2ZXV
 
 | | Crystal structure of putative acetyltransferase from T. thermophilus HB8 | | Descriptor: | Putative uncharacterized protein TTHA1799 | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Genetic Encoding of 3-Iodo-l-Tyrosine in Escherichia coli for Single-Wavelength Anomalous Dispersion Phasing in Protein Crystallography
Structure, 17, 2009
|
|
1YMW
 
 | | The study of reductive unfolding pathways of RNase A (Y92G mutant) | | Descriptor: | Ribonuclease pancreatic | | Authors: | Xu, G, Narayan, M, Kurinov, I, Ripoll, D.R, Welker, E, Khalili, M, Ealick, S.E, Scheraga, H.A. | | Deposit date: | 2005-01-21 | | Release date: | 2006-01-31 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A localized specific interaction alters the unfolding pathways of structural homologues.
J.Am.Chem.Soc., 128, 2006
|
|
1YMR
 
 | | The study of reductive unfolding pathways of RNase A (Y92A mutant) | | Descriptor: | Ribonuclease pancreatic | | Authors: | Xu, G, Narayan, M, Kurinov, I, Ripoll, D.R, Welker, E, Khalili, M, Ealick, S.E, Scheraga, H.A. | | Deposit date: | 2005-01-21 | | Release date: | 2006-01-31 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A localized specific interaction alters the unfolding pathways of structural homologues.
J.Am.Chem.Soc., 128, 2006
|
|
1YMN
 
 | | The study of reductive unfolding pathways of RNase A (Y92L mutant) | | Descriptor: | Ribonuclease pancreatic | | Authors: | Xu, G, Narayan, M, Kurinov, I, Ripoll, D.R, Welker, E, Khalili, M, Ealick, S.E, Scheraga, H.A. | | Deposit date: | 2005-01-21 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A localized specific interaction alters the unfolding pathways of structural homologues.
J.Am.Chem.Soc., 128, 2006
|
|
