4WJN
 
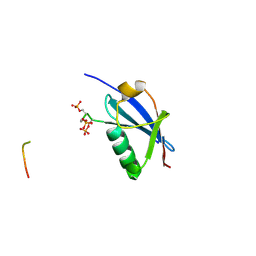 | | Crystal structure of SUMO1 in complex with phosphorylated PML | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Cappadocia, L, Mascle, X.H, Bourdeau, V, Tremblay-Belzile, S, Chaker-Margot, M, Lussier-Price, M, Wada, J, Sakaguchi, K, Aubry, M, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of the Phosphorylation-Dependent Interaction between PML and SUMO1.
Structure, 23, 2015
|
|
4WJP
 
 | | Crystal Structure of SUMO1 in complex with phosphorylated Daxx | | Descriptor: | Daxx, Small ubiquitin-related modifier 1 | | Authors: | Cappadocia, L, Mascle, X.H, Bourdeau, V, Tremblay-Belzile, S, Chaker-Margot, M, Lussier-Price, M, Wada, J, Sakaguchi, K, Aubry, M, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characterization of the Phosphorylation-Dependent Interaction between PML and SUMO1.
Structure, 23, 2015
|
|
4WJO
 
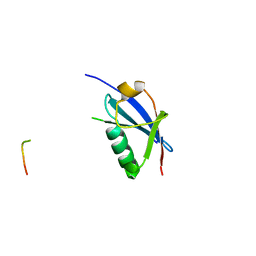 | | Crystal Structure of SUMO1 in complex with PML | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Cappadocia, L, Mascle, X.H, Bourdeau, V, Tremblay-Belzile, S, Chaker-Margot, M, Lussier-Price, M, Wada, J, Sakaguchi, K, Aubry, M, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural and Functional Characterization of the Phosphorylation-Dependent Interaction between PML and SUMO1.
Structure, 23, 2015
|
|
5SXP
 
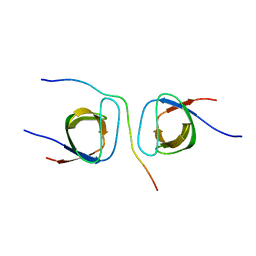 | | STRUCTURAL BASIS FOR THE INTERACTION BETWEEN ITCH PRR AND BETA-PIX | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog, Rho guanine nucleotide exchange factor 7 | | Authors: | Cappadocia, L, Desrochers, G, Lussier-Price, M, Angers, A, Omichinski, J.G. | | Deposit date: | 2016-08-09 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis of interactions between SH3 domain-containing proteins and the proline-rich region of the ubiquitin ligase Itch.
J. Biol. Chem., 292, 2017
|
|
5C17
 
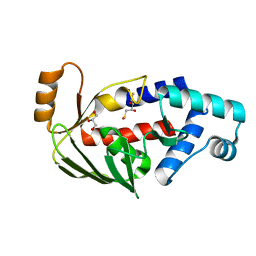 | | Crystal structure of the mercury-bound form of MerB2 | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-13 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
5C0U
 
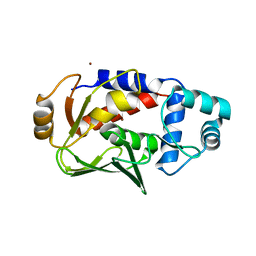 | | Crystal structure of the copper-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, COPPER (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
5C0T
 
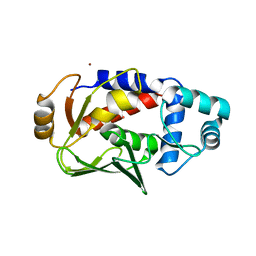 | | Crystal structure of the mercury-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
5DSF
 
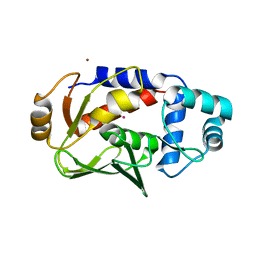 | | Crystal structure of the mercury-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-09-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
8T4T
 
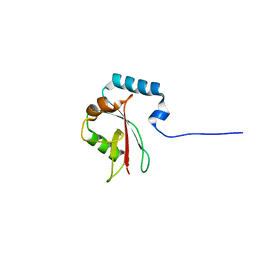 | |
8T31
 
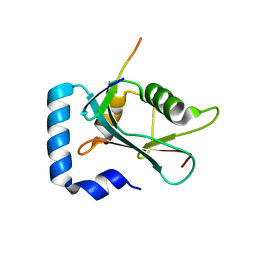 | |
8T35
 
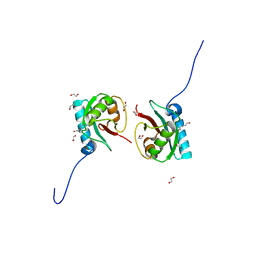 | | Crystal structure of K51 acetylated LC3A in complex with the LIR of TP53INP2/DOR | | Descriptor: | 1,2-ETHANEDIOL, Tumor protein p53-inducible nuclear protein 2,Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 2024
|
|
8T33
 
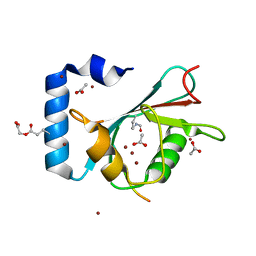 | | Crystal structure of K46 acetylated GABARAP in complex with the LIR of TP53INP2/DOR | | Descriptor: | ACETATE ION, Gamma-aminobutyric acid receptor-associated protein, Tumor protein p53-inducible nuclear protein 2, ... | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 2024
|
|
8T32
 
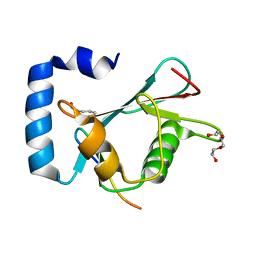 | | Crystal structure of K48 acetylated GABARAP in complex with the LIR of TP53INP2/DOR | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein, LIR of DOR, TRIETHYLENE GLYCOL | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 2024
|
|
1DL6
 
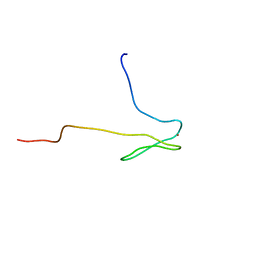 | | SOLUTION STRUCTURE OF HUMAN TFIIB N-TERMINAL DOMAIN | | Descriptor: | TRANSCRIPTION FACTOR II B (TFIIB), ZINC ION | | Authors: | Chen, H.-T, Legault, P, Glushka, J, Omichinski, J.G, Scott, R.A. | | Deposit date: | 1999-12-08 | | Release date: | 2000-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a (Cys3His) zinc ribbon, a ubiquitous motif in archaeal and eucaryal transcription.
Protein Sci., 9, 2000
|
|
1OLH
 
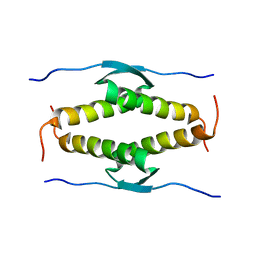 | |
8T36
 
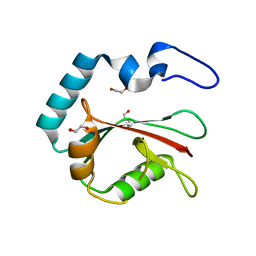 | | Crystal structure of K49 acetylated LC3A in complex with the LIR of TP53INP2/DOR | | Descriptor: | 1,2-ETHANEDIOL, Tumor protein p53-inducible nuclear protein 2,Microtubule-associated proteins 1A/1B light chain 3A chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 2024
|
|
3F2F
 
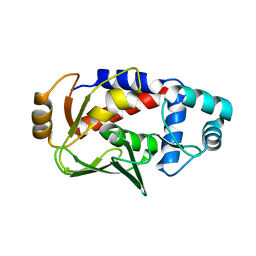 | | Crystal structure of the mercury-bound form of MerB, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3F0P
 
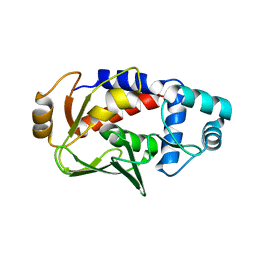 | | Crystal structure of the mercury-bound form of MerB, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-25 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3F0O
 
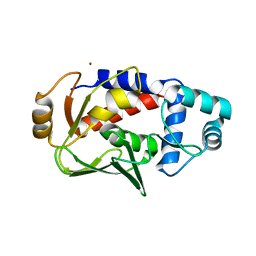 | | Crystal structure of MerB, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase, BROMIDE ION | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-25 | | Release date: | 2008-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3F2H
 
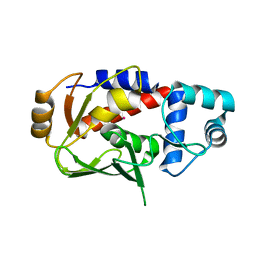 | | Crystal structure of the mercury-bound form of MerB mutant C160S, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase, MERCURY (II) ION | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
3F2G
 
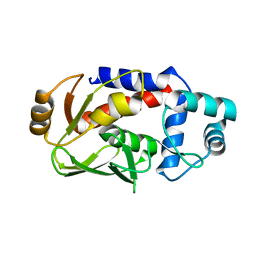 | | Crystal structure of MerB mutant C160S, the Organomercurial Lyase involved in a bacterial mercury resistance system | | Descriptor: | Alkylmercury lyase | | Authors: | Lafrance-Vanasse, J, Lefebvre, M, Di Lello, P, Sygusch, J, Omichinski, J.G. | | Deposit date: | 2008-10-29 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
J.Biol.Chem., 284, 2009
|
|
5U7A
 
 | | Crystal structure of a complex formed between MerB and Dimethyltin | | Descriptor: | Alkylmercury lyase, BROMIDE ION, Dimethyltin dibromide, ... | | Authors: | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.532 Å) | | Cite: | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5U79
 
 | | Crystal structure of a complex formed between MerB and Dimethyltin | | Descriptor: | ACETATE ION, Alkylmercury lyase, BROMIDE ION, ... | | Authors: | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5U7C
 
 | | Crystal structure of the lead-bound form of MerB formed from diethyllead. | | Descriptor: | ACETATE ION, Alkylmercury lyase, BROMIDE ION, ... | | Authors: | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
5U82
 
 | | Crystal structure of a MerB-triethyltin complex | | Descriptor: | ACETATE ION, Alkylmercury lyase, BROMIDE ION, ... | | Authors: | Wahba, H.M, Stevenson, M, Mansour, A, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2016-12-13 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural and Biochemical Characterization of Organotin and Organolead Compounds Binding to the Organomercurial Lyase MerB Provide New Insights into Its Mechanism of Carbon-Metal Bond Cleavage.
J. Am. Chem. Soc., 139, 2017
|
|
