6ORC
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | Sel1 repeat protein | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-29 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
7SF6
 
 | | Crystal Structure of Siderophore Binding Protein FatB from Desulfitobacterium hafniense | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,3-DIHYDROXY-BENZOYLAMINO)-3-HYDROXY-PROPIONIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Patel, H.P, Nordquist, K.A, Schaab, K.M, Sha, J, Babnigg, G, Bond, A.H, Joachimiak, A, Midwest Center for Structural Genomics, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2021-10-03 | | Release date: | 2021-12-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of Siderophore Binding Protein FatB from Desulfitobacterium hafniense
To Be Published
|
|
5L1A
 
 | | Crystal structure of uncharacterized protein LPG2271 from Legionella pneumophila | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Uncharacterized protein | | Authors: | Chang, C, Xu, X, Cui, H, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-07-28 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of uncharacterized protein LPG2271 from Legionella pneumophila
To Be Published
|
|
5L10
 
 | | Crystal Structure of N-Acylhomoserine Lactone Dependent LuxR Family Transcriptionl Factor CepR2 from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winan, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-07-28 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of N-Acylhomoserine Lactone Dependent LuxR Family Transcriptionl Factor CepR2 from Burkholderia cenocepacia.
To Be Published
|
|
6NJC
 
 | | Crystal Structure of the Sialate O-acetylesterase from Bacteroides vulgatus | | Descriptor: | ACETIC ACID, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Li, H, Biglow, L, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-01-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Sialate O-acetylesterase from Bacteroides vulgatus
To Be Published
|
|
6OK0
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Sel1 repeat protein, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
4XEA
 
 | | Crystal structure of putative M16-like peptidase from Alicyclobacillus acidocaldarius | | Descriptor: | ACETATE ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Michalska, K, Tesar, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-23 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of putative M16-like peptidase from Alicyclobacillus acidocaldarius
To Be Published
|
|
4XED
 
 | | PKD domain of M14-like peptidase from Thermoplasmatales archaeon SCGC AB-540-F20 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Peptidase M14, ... | | Authors: | Michalska, K, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PKD domain of M14-like peptidase from Thermoplasmatales archaeon SCGC AB-540-F20
To Be Published
|
|
4XXT
 
 | | Crystal structure of Fused Zn-dependent amidase/peptidase/peptodoglycan-binding domain-containing protein from Clostridium acetobutylicum ATCC 824 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Fusion of predicted Zn-dependent amidase/peptidase (Cell wall hydrolase/DD-carboxypeptidase family) and uncharacterized domain of ErfK family peptodoglycan-binding domain, ... | | Authors: | Chang, C, Cuff, M, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-30 | | Release date: | 2015-02-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Fused Zn-dependent amidase/peptidase/peptodoglycan-binding domain-containing protein from from Clostridium acetobutylicum ATCC 824
To Be Published
|
|
4YCS
 
 | | Crystal structure of putative lipoprotein from Peptoclostridium difficile 630 (fragment) | | Descriptor: | ACETATE ION, GLYCEROL, SODIUM ION, ... | | Authors: | Michalska, K, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-20 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of putative lipoprotein from Peptoclostridium difficile 630 (fragment)
To Be Published
|
|
4WHI
 
 | | Crystal structure of C-terminal domain of penicillin binding protein Rv0907 | | Descriptor: | BROMIDE ION, Beta-lactamase, NICKEL (II) ION | | Authors: | Chang, C, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-22 | | Release date: | 2014-10-08 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of C-terminal domain of penicillin binding protein Rv0907
To Be Published
|
|
4XVO
 
 | | L,D-transpeptidase from Mycobacterium smegmatis | | Descriptor: | L,D-transpeptidase, PHOSPHATE ION | | Authors: | Osipiuk, J, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-27 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | L,D-transpeptidase from Mycobacterium smegmatis
to be published
|
|
4XS5
 
 | | Crystal structure of Sulfate transporter/antisigma-factor antagonist STAS from Dyadobacter fermentans DSM 18053 | | Descriptor: | Sulfate transporter/antisigma-factor antagonist STAS | | Authors: | Chang, C, Cuff, M, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-21 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Sulfate transporter/antisigma-factor antagonist STAS from Dyadobacter fermentans DSM 18053
To Be Published
|
|
4YF1
 
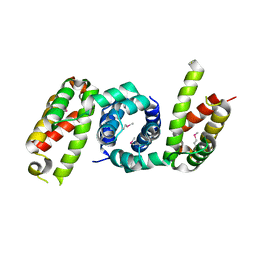 | | 1.85 angstrom crystal structure of lmo0812 from Listeria monocytogenes EGD-e | | Descriptor: | CITRATE ANION, Lmo0812 protein, SODIUM ION | | Authors: | Krishna, S.N, Light, S.H, Filippova, E.V, Minasov, G, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 angstrom crystal structure of lmo0812 from Listeria monocytogenes EGD-e
To Be Published
|
|
5HX0
 
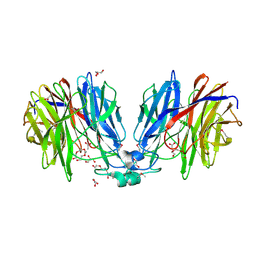 | | Crystal structure of unknown function protein Dfer_1899 fromDyadobacter fermentans DSM 18053 | | Descriptor: | ACETATE ION, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Chang, C, Duke, N, Clancy, S, Chhor, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-01-29 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Crystal structure of unknown function protein Dfer_1899 fromDyadobacter fermentans DSM 18053
To Be Published
|
|
5JBR
 
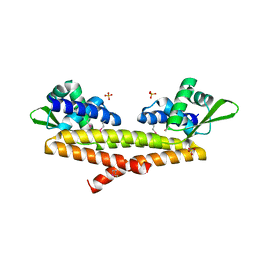 | | Crystal structure of uncharacterized protein Bcav_2135 from Beutenbergia cavernae | | Descriptor: | SULFATE ION, Uncharacterized protein Bcav_2135 | | Authors: | Chang, C, Cuff, M, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-04-13 | | Release date: | 2016-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of uncharacterized protein Bcav_2135 from Beutenbergia cavernae
To Be Published
|
|
7LZG
 
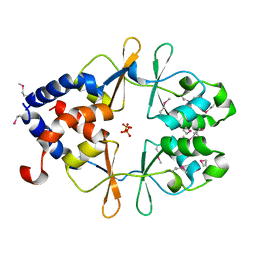 | |
5I2H
 
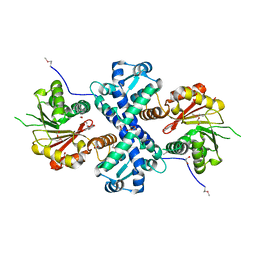 | | Crystal structure of O-methyltransferase family 2 protein Plim_1147 from Planctomyces limnophilus DSM 3776 complex with Apigenin | | Descriptor: | 1,2-ETHANEDIOL, 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, FORMIC ACID, ... | | Authors: | Chang, C, Duke, N, Bigelow, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-02-08 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Crystal structure of O-methyltransferase family 2 protein Plim_1147 from Planctomyces limnophilus DSM 3776 complex with Apigenin.
To Be Published
|
|
5DS0
 
 | | Crystal structure of TET aminopeptidase from marine sediment archaeon Thaumarchaeota archaeon SCGC AB-539-E09 | | Descriptor: | COBALT (II) ION, GLYCEROL, Peptidase M42 | | Authors: | Michalska, K, Chhor, G, Mootz, J, Endres, M, Jedrzejczak, R, Babnigg, G, Steen, A, Lloyd, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-09-16 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of TET aminopeptidase from marine sediment archaeon Thaumarchaeota archaeon SCGC AB-539-E09
To Be Published
|
|
5ES2
 
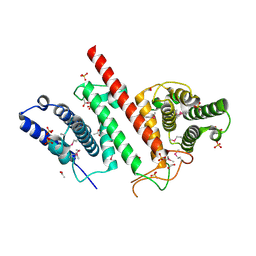 | | The crystal structure of a functionally uncharacterized protein LPG0634 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-11-16 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of a functionally uncharacterized protein LPG0634 from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To Be Published
|
|
5DYF
 
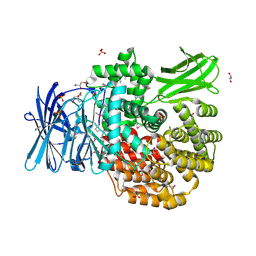 | | The crystal structure of Aminopeptidase N in complex with N-benzyl-1,2-diaminoethylphosphonic acid | | Descriptor: | Aminopeptidase N, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nocek, B, Joachimiak, A, Vassiliou, S, Berlicki, L, Mucha, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-09-24 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | The crystal structure of Aminopeptidase N in complex with N-benzyl-1,2-diaminoethylphosphonic acid
To Be Published
|
|
5DU2
 
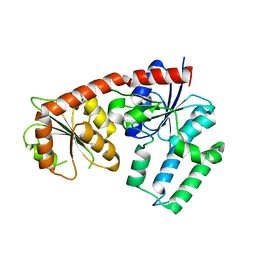 | | Structural analysis of EspG2 glycosyltransferase | | Descriptor: | EspG2 glycosyltransferase | | Authors: | Michalska, K, Elshahawi, S.I, Bigelow, L, Babnigg, G, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-09-18 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of EspG2 glycosyltransferase
To Be Published
|
|
5EVI
 
 | | Crystal Structure of Beta-Lactamase/D-Alanine Carboxypeptidase from Pseudomonas syringae | | Descriptor: | 1,2-ETHANEDIOL, Beta-Lactamase/D-Alanine Carboxypeptidase, SULFATE ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-11-19 | | Release date: | 2016-01-13 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Beta-Lactamase/D-Alanine Carboxypeptidase from Pseudomonas syringae
To Be Published
|
|
5I47
 
 | | Crystal structure of RimK domain protein ATP-grasp from Sphaerobacter thermophilus DSM 20745 | | Descriptor: | GLYCEROL, RimK domain protein ATP-grasp | | Authors: | Chang, C, Duke, N, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-02-11 | | Release date: | 2016-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of RimK domain protein ATP-grasp from Sphaerobacter thermophilus DSM 20745
To Be Published
|
|
6VEK
 
 | | Contact-dependent growth inhibition toxin-immunity protein complex from from E. coli 3006, full-length | | Descriptor: | contact-dependent immunity protein CdiI, contact-dependent toxin CdiA | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-01-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Contact-dependent growth inhibition toxin-immunity protein complex from from E. coli 3006, full-length
To Be Published
|
|
