8FX3
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to Immucillin-GP, showing the structure of the complete active site in its open conformation | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, PHOSPHORIC ACID MONO-[5-(2-AMINO-4-OXO-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-3,4-DIHYDROXY-PYRROLIDIN-2-YLMETHYL] ESTER | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Suthagar, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX1
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to (R)-SerMe-ImmH Phosphonate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, [(3R)-4-hydroxy-3-{[(4-oxo-4,5-dihydro-3H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]amino}butyl]phosphonic acid | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Clinch, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX0
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to (S)-SerMe-ImmH Phosphonate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, [(3R)-4-hydroxy-3-{[(4-oxo-4,5-dihydro-3H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]amino}butyl]phosphonic acid | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Clinch, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FX2
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to Immucillin-HP | | Descriptor: | (1S)-1(9-DEAZAHYPOXANTHIN-9YL)1,4-DIDEOXY-1,4-IMINO-D-RIBITOL-5-PHOSPHATE, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Suthagar, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
8FWZ
 
 | | Crystal structure of the Trypanosoma cruzi hypoxanthine-guanine-xanthine phosphoribosyltransferase (HGXPRT), isoform D, bound to Hydroxypropyl-Lin-ImmH Phosphonate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, PHOSPHATE ION, [(2R)-2-hydroxy-3-{[(4-oxo-4,5-dihydro-3H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]amino}propyl]phosphonic acid | | Authors: | Hughes, R, Meneely, K.M, Glockzin, K, Clinch, K, Tyler, P.C, Lamb, A.L, Meek, T.D, Katzfuss, A. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Kinetic and Structural Characterization of Trypanosoma cruzi Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferases and Repurposing of Transition-State Analogue Inhibitors.
Biochemistry, 62, 2023
|
|
7UEZ
 
 | | Apo RibB from Vibrio cholera | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase | | Authors: | Kenjic, N, Meneely, K.M, Lamb, A.L. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Evidence for the Chemical Mechanism of RibB (3,4-Dihydroxy-2-butanone 4-phosphate Synthase) of Riboflavin Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
7UF5
 
 | | RibB from Vibrio cholera bound with intermediate 2 in the reaction cycle and the products DHBP and formate | | Descriptor: | (2R)-2-(dihydroxymethyl)-2-hydroxy-3-oxobutyl dihydrogen phosphate, (2R)-2-hydroxy-3-oxobutyl dihydrogen phosphate, 3,4-dihydroxy-2-butanone 4-phosphate synthase, ... | | Authors: | Kenjic, N, Meneely, K.M, Lamb, A.L. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evidence for the Chemical Mechanism of RibB (3,4-Dihydroxy-2-butanone 4-phosphate Synthase) of Riboflavin Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
7UF0
 
 | | RibB from Vibrio cholera bound with D-ribulose-5-phosphate (D-Ru5P) | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, RIBULOSE-5-PHOSPHATE | | Authors: | Kenjic, N, Meneely, K.M, Lamb, A.L. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for the Chemical Mechanism of RibB (3,4-Dihydroxy-2-butanone 4-phosphate Synthase) of Riboflavin Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
7UF1
 
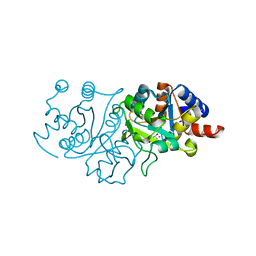 | | RibB from Vibrio cholera bound with D-Ribose-5-phosphate (D-R5P) and manganese | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, MANGANESE (II) ION, RIBOSE-5-PHOSPHATE | | Authors: | Kenjic, N, Meneely, K.M, Lamb, A.L. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evidence for the Chemical Mechanism of RibB (3,4-Dihydroxy-2-butanone 4-phosphate Synthase) of Riboflavin Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
7UF4
 
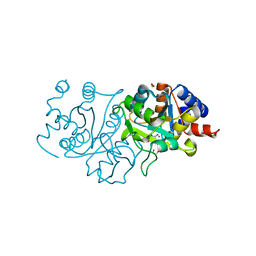 | | RibB from Vibrio cholera bound with intermediate 1 of the reaction cycle and D-ribulose-5-phosphate (D-Ru5P) | | Descriptor: | (2R)-2-hydroxy-3,4-dioxopentyl dihydrogen phosphate, 1,2-ETHANEDIOL, 3,4-dihydroxy-2-butanone 4-phosphate synthase, ... | | Authors: | Kenjic, N, Meneely, K.M, Lamb, A.L. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evidence for the Chemical Mechanism of RibB (3,4-Dihydroxy-2-butanone 4-phosphate Synthase) of Riboflavin Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
7UF2
 
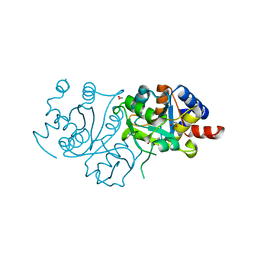 | | RibB from Vibrio cholera bound with D-xylulose-5-phosphate (D-Xy5P) and manganese | | Descriptor: | 1,2-ETHANEDIOL, 3,4-dihydroxy-2-butanone 4-phosphate synthase, 5-O-phosphono-D-xylulose, ... | | Authors: | Kenjic, N, Meneely, K.M, Lamb, A.L. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evidence for the Chemical Mechanism of RibB (3,4-Dihydroxy-2-butanone 4-phosphate Synthase) of Riboflavin Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
7UF3
 
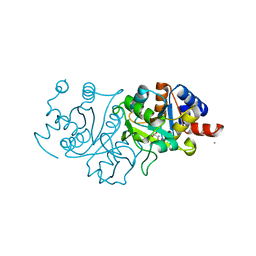 | | RibB from Vibrio cholera bound with L-xylulose-5-phosphate (L-Xy5P) and manganese | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, L-xylulose-5-phosphate, MANGANESE (II) ION | | Authors: | Kenjic, N, Meneely, K.M, Lamb, A.L. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evidence for the Chemical Mechanism of RibB (3,4-Dihydroxy-2-butanone 4-phosphate Synthase) of Riboflavin Biosynthesis.
J.Am.Chem.Soc., 144, 2022
|
|
3E6I
 
 | |
6NN4
 
 | | The structure of human liver pyruvate kinase, hLPYK-D499N, in complex with Fru-1,6-BP | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, PHOSPHOENOLPYRUVATE, ... | | Authors: | McFarlane, J.S, Ronnebaum, T.A, Meneely, K.M, Fenton, A.W, Lamb, A.L. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Changes in the allosteric site of human liver pyruvate kinase upon activator binding include the breakage of an intersubunit cation-pi bond.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6NN5
 
 | | The structure of human liver pyruvate kinase, hLPYK-W527H | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pyruvate kinase PKLR | | Authors: | McFarlane, J.S, Ronnebaum, T.A, Meneely, K.M, Fenton, A.W, Lamb, A.L. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.256 Å) | | Cite: | Changes in the allosteric site of human liver pyruvate kinase upon activator binding include the breakage of an intersubunit cation-pi bond.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6NN7
 
 | | The structure of human liver pyruvate kinase, hLPYK-GGG | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, GLYCEROL, ... | | Authors: | McFarlane, J.S, Ronnebaum, T.A, Meneely, K.M, Fenton, A.W, Lamb, A.L. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Changes in the allosteric site of human liver pyruvate kinase upon activator binding include the breakage of an intersubunit cation-pi bond.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6NN8
 
 | | The structure of human liver pyruvate kinase, hLPYK-S531E | | Descriptor: | 1,2-ETHANEDIOL, Pyruvate kinase PKLR | | Authors: | McFarlane, J.S, Ronnebaum, T.A, Meneely, K.M, Fenton, A.W, Lamb, A.L. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | Changes in the allosteric site of human liver pyruvate kinase upon activator binding include the breakage of an intersubunit cation-pi bond.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
3S5W
 
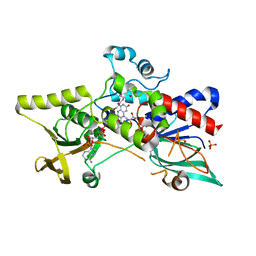 | | Ornithine Hydroxylase (PvdA) from Pseudomonas aeruginosa | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-ornithine 5-monooxygenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Olucha, J, Lamb, A.L. | | Deposit date: | 2011-05-23 | | Release date: | 2011-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two Structures of an N-Hydroxylating Flavoprotein Monooxygenase: ORNITHINE HYDROXYLASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 286, 2011
|
|
3S61
 
 | |
3T3S
 
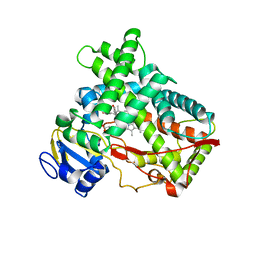 | | Human Cytochrome P450 2A13 in complex with Pilocarpine | | Descriptor: | (3S,4R)-3-ethyl-4-[(1-methyl-1H-imidazol-5-yl)methyl]dihydrofuran-2(3H)-one, Cytochrome P450 2A13, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | DeVore, N.M, Scott, E.E. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine.
Febs J., 279, 2012
|
|
3T3R
 
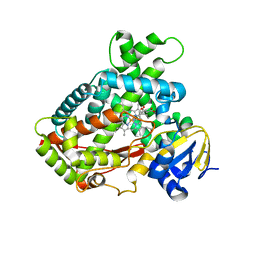 | | Human Cytochrome P450 2A6 in complex with Pilocarpine | | Descriptor: | (3S,4R)-3-ethyl-4-[(1-methyl-1H-imidazol-5-yl)methyl]dihydrofuran-2(3H)-one, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | DeVore, N.M, Scott, E.E. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine.
Febs J., 279, 2012
|
|
3T3Q
 
 | | Human Cytochrome P450 2A6 I208S/I300F/G301A/S369G in complex with Pilocarpine | | Descriptor: | (3S,4R)-3-ethyl-4-[(1-methyl-1H-imidazol-5-yl)methyl]dihydrofuran-2(3H)-one, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | DeVore, N.M, Scott, E.E. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine.
Febs J., 279, 2012
|
|
5KRQ
 
 | | Renalase in complex with NADPH | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Renalase | | Authors: | Silvaggi, N.R, Moran, G.R, Roman, J.V. | | Deposit date: | 2016-07-07 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.086 Å) | | Cite: | Ligand binding phenomena that pertain to the metabolic function of renalase.
Arch.Biochem.Biophys., 612, 2016
|
|
