3C1T
 
 | | Binding of two substrate analogue molecules to dihydroflavonol 4-reductase alters the functional geometry of the catalytic site | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, dihydroflavonol 4-reductase | | Authors: | Trabelsi, N, Petit, P, Granier, T, Langlois d'Estaintot, B, Delrot, S, Gallois, B. | | Deposit date: | 2008-01-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Structural evidence for the inhibition of grape dihydroflavonol 4-reductase by flavonols
Acta Crystallogr.,Sect.D, D64, 2008
|
|
5LVS
 
 | | Self-assembled protein-aromatic foldamer complexes with 2:3 and 2:2:1 stoichiometries | | Descriptor: | Aromatic foldamer, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Jewginski, M, LANGLOIS D'ESTAINTOT, B, Granier, T, Huc, Y. | | Deposit date: | 2016-09-14 | | Release date: | 2017-03-08 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Self-Assembled Protein-Aromatic Foldamer Complexes with 2:3 and 2:2:1 Stoichiometries.
J. Am. Chem. Soc., 139, 2017
|
|
7PCJ
 
 | | X-ray structure of CypA-C52AK125C/CsA/aromatic foldamer complex | | Descriptor: | Aromatic foldamer, Cyclosporin A, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Vallade, M, Langlois d'Estaintot, B, Fischer, L, Buratto, J, Savko, M, Huc, I. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | X-ray structure of a cystein mutant of Cyclophilin A tethered to an aromatic oligoamide foldamer complexed with Cyclosporin A
To Be Published
|
|
7QBW
 
 | |
4MKR
 
 | | Structure of the apo form of a Zingiber officinale double bond reductase | | Descriptor: | Zingiber officinale double bond reductase | | Authors: | Buratto, J, Langlois d'Estaintot, B, Granier, T, Gallois, B, Willis, M.A, Sang, Y, Flores-Sanchez, I.J, Gang, D.R. | | Deposit date: | 2013-09-05 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of Zingiber officinale double bond reductase
To be Published
|
|
198D
 
 | | A TRIGONAL FORM OF THE IDARUBICIN-D(CGATCG) COMPLEX: CRYSTAL AND MOLECULAR STRUCTURE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), IDARUBICIN, SPERMINE | | Authors: | Dautant, A, Langlois D'Estaintot, B, Gallois, B, Brown, T, Hunter, W.N. | | Deposit date: | 1994-11-28 | | Release date: | 1995-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A trigonal form of the idarubicin:d(CGATCG) complex; crystal and molecular structure at 2.0 A resolution.
Nucleic Acids Res., 23, 1995
|
|
3HFS
 
 | | Structure of apo anthocyanidin reductase from vitis vinifera | | Descriptor: | Anthocyanidin reductase, CHLORIDE ION | | Authors: | Gargouri, M, Mauge, C, Langlois d'Estaintot, B, Granier, T, Manigan, C, Gallois, B. | | Deposit date: | 2009-05-12 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structure and epimerase activity of anthocyanidin reductase from Vitis vinifera.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4MTY
 
 | | Structure at 1A resolution of a helical aromatic foldamer-protein complex. | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ogayone, T, Buratto, J, Langlois D'Estaintot, B, Stupfel, M, Granier, T, Gallois, B, Huc, Y. | | Deposit date: | 2013-09-20 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of a complex formed by a protein and a helical aromatic oligoamide foldamer at 2.1 a resolution.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1KRQ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CAMPYLOBACTER JEJUNI FERRITIN | | Descriptor: | ferritin | | Authors: | Hortolan, L, Saintout, N, Granier, G, Langlois d'Estaintot, B, Manigand, C, Mizunoe, Y, Wai, S.N, Gallois, B, Precigoux, G. | | Deposit date: | 2002-01-10 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | STRUCTURE OF CAMPYLOBACTER JEJUNI FERRITIN AT 2.7 A RESOLUTION
To be Published
|
|
482D
 
 | | RELEASE OF THE CYANO MOIETY IN THE CRYSTAL STRUCTURE OF N-CYANOMETHYL-N-(2-METHOXYETHYL)-DAUNOMYCIN COMPLEXED WITH D(CGATCG) | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*G)-3', N-HYDROXYMETHYL-N-(2-METHOXYETHYL)-DAUNOMYCIN | | Authors: | Saminadin, P, Dautant, A, Mondon, M, Langlois D'Estaintot, B, Courseille, C, Precigoux, G. | | Deposit date: | 1999-07-27 | | Release date: | 1999-09-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Release of the cyano moiety in the crystal structure of N-cyanomethyl-N-(2-methoxyethyl)-daunomycin complexed with d(CGATCG).
Eur.J.Biochem., 267, 2000
|
|
1LB3
 
 | | Structure of recombinant mouse L chain ferritin at 1.2 A resolution | | Descriptor: | CADMIUM ION, FERRITIN LIGHT CHAIN 1, GLYCEROL, ... | | Authors: | Granier, T, Langlois D'Estaintot, B, Gallois, B, Chevalier, J.-M, Precigoux, G, Santambrogio, P, Arosio, P. | | Deposit date: | 2002-04-02 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural description of the active sites of mouse L-chain ferritin at 1.2A resolution
J.Biol.Inorg.Chem., 8, 2003
|
|
1D82
 
 | |
2IOD
 
 | | Binding of two substrate analogue molecules to dihydroflavonol-4-reductase alters the functional geometry of the catalytic site | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Dihydroflavonol 4-reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petit, P, Langlois d'Estaintot, B, Granier, T, Gallois, B. | | Deposit date: | 2006-10-10 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Binding of two substrate analogue molecules to dihydroflavonol-4-reductase alters the functional geometry of the catalytic site
To be Published
|
|
3CKU
 
 | |
2IBA
 
 | | Urate oxidase from Aspergillus flavus complexed with its inhibitor 8-azaxanthine | | Descriptor: | 8-AZAXANTHINE, SODIUM ION, Uricase | | Authors: | Colloc'h, N, Retailleau, P, Sopkova-de Oliveira Santos, J, Prange, T. | | Deposit date: | 2006-09-11 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein Crystallography under Xenon and Nitrous Oxide Pressure: Comparison with In Vivo Pharmacology Studies and Implications for the Mechanism of Inhaled Anesthetic Action
Biophys.J., 92, 2007
|
|
2ICQ
 
 | | urate oxidase under 2.0 MPa pressure of nitrous oxide | | Descriptor: | 8-AZAXANTHINE, CYSTEINE, NITROUS OXIDE, ... | | Authors: | Colloc'h, N, Sopkova-de Oliveira Santos, J, Retailleau, P, Prange, T, Abraini, J.H. | | Deposit date: | 2006-09-13 | | Release date: | 2007-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein Crystallography under Xenon and Nitrous Oxide Pressure: Comparison with In Vivo Pharmacology Studies and Implications for the Mechanism of Inhaled Anesthetic Action
Biophys.J., 92, 2007
|
|
2IC0
 
 | | Urate oxidase under 2.0 MPa pressure of xenon | | Descriptor: | 8-AZAXANTHINE, SODIUM ION, Uricase, ... | | Authors: | Colloc'h, N, Retailleau, P, Sopkova-de Oliveira Santos, J, Abraini, J.H, Prange, T. | | Deposit date: | 2006-09-12 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Protein Crystallography under Xenon and Nitrous Oxide Pressure: Comparison with In Vivo Pharmacology Studies and Implications for the Mechanism of Inhaled Anesthetic Action
Biophys.J., 92, 2007
|
|
5MY3
 
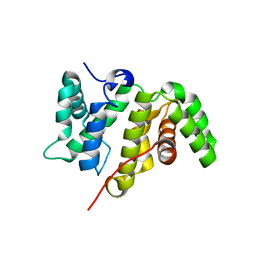 | | Crystal structure of the RhoGAP domain of Rgd1p at 2.19 Angstroms resolution | | Descriptor: | RHO GTPase-activating protein RGD1 | | Authors: | Martinez, D.M, d'Estaintot, B.L, Granier, T, Hugues, M, Odaert, B, Gallois, B, Doignon, F. | | Deposit date: | 2017-01-25 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural evidence of a phosphoinositide-binding site in the Rgd1-RhoGAP domain.
Biochem. J., 474, 2017
|
|
1DAT
 
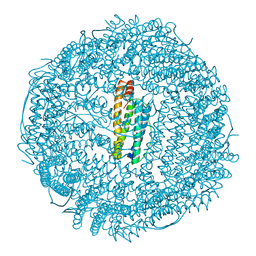 | | CUBIC CRYSTAL STRUCTURE RECOMBINANT HORSE L APOFERRITIN | | Descriptor: | CADMIUM ION, L FERRITIN | | Authors: | Gallois, B, Granier, T, Langlois D'Estaintot, B, Crichton, R.R, Roland, F. | | Deposit date: | 1996-11-14 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray structure of recombinant horse L-chain apoferritin at 2.0 angstrom resolution: Implications for stability and function.
J.Biol.Inorg.Chem., 2, 1997
|
|
1D67
 
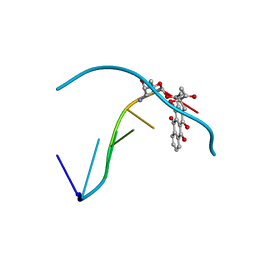 | | THE MOLECULAR STRUCTURE OF AN IDARUBICIN-D(TGATCA) COMPLEX AT HIGH RESOLUTION | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*CP*A)-3'), IDARUBICIN | | Authors: | Gallois, B, Langlois D'Estaintot, B, Brown, T, Hunter, W.N. | | Deposit date: | 1992-03-31 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of an idarubicin-d(TGATCA) complex at high resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
6Q3O
 
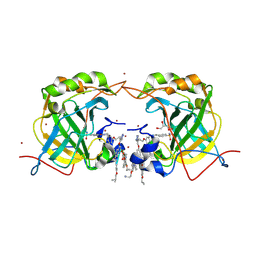 | | PROTEIN-AROMATIC FOLDAMER COMPLEX CRYSTAL STRUCTURE | | Descriptor: | 4-sulfamoylbenzoic acid, Aromatic foldamer, Carbonic anhydrase 2, ... | | Authors: | Zeberko, C, Langlois d'Estaintot, B, Fischer, L, Granier, T, Kauffmann, B, Huc, I. | | Deposit date: | 2018-12-04 | | Release date: | 2019-12-25 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | PROTEIN-AROMATIC FOLDAMER COMPLEX CRYSTAL STRUCTURE
To Be Published
|
|
1HRS
 
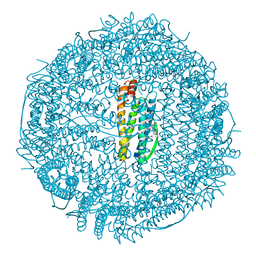 | | A CRYSTALLOGRAPHIC STUDY OF HAEM BINDING TO FERRITIN | | Descriptor: | APOFERRITIN CO-CRYSTALLIZED WITH SN-PROTOPORPHYRIN IX IN CADMIUM SULFATE, CADMIUM ION, PROTOPORPHYRIN IX | | Authors: | Precigoux, G, Yariv, J, Gallois, B, Dautant, A, Courseille, C, Langlois D'Estaintot, B. | | Deposit date: | 1993-11-05 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A crystallographic study of haem binding to ferritin.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1DA0
 
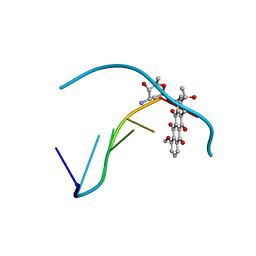 | | DNA-DRUG INTERACTIONS: THE CRYSTAL STRUCTURE OF D(CGATCG) COMPLEXED WITH DAUNOMYCIN | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3') | | Authors: | Moore, M.H, Hunter, W.N, Langlois D'Estaintot, B, Kennard, O. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | DNA-drug interactions. The crystal structure of d(CGATCG) complexed with daunomycin.
J.Mol.Biol., 206, 1989
|
|
2H0K
 
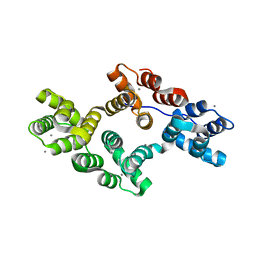 | | Crystal Structure of a Mutant of Rat Annexin A5 | | Descriptor: | Annexin A5, CALCIUM ION | | Authors: | Granier, T, Langlois D'Estaintot, B, Gallois, B, Tessier, B, Brisson, A. | | Deposit date: | 2006-05-15 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Annexin-A5 assembled into two-dimensional arrays promotes cell membrane repair.
Nat Commun, 2, 2011
|
|
184D
 
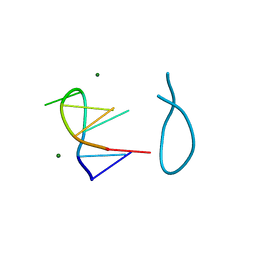 | | SELF-ASSOCIATION OF A DNA LOOP CREATES A QUADRUPLEX: CRYSTAL STRUCTURE OF D(GCATGCT) AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3'), MAGNESIUM ION | | Authors: | Leonard, G.A, Zhang, S, Peterson, M.R, Harrop, S.J, Helliwell, J.R, Cruse, W.B.T, Langlois D'Estaintot, B, Kennard, O, Brown, T, Hunter, W.N. | | Deposit date: | 1994-08-10 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Self-association of a DNA loop creates a quadruplex: crystal structure of d(GCATGCT) at 1.8 A resolution.
Structure, 3, 1995
|
|
