3BNU
 
 | | Crystal structure of polyamine oxidase FMS1 from Saccharomyces cerevisiae in complex with bis-(3S,3'S)-methylated spermine | | Descriptor: | (3S,3'S)-N~1~,N~1~'-butane-1,4-diyldibutane-1,3-diamine, FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1 | | Authors: | Huang, Q, Hao, Q. | | Deposit date: | 2007-12-14 | | Release date: | 2008-01-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the substrate stereospecificity of FMS1.
To Be Published
|
|
3CNP
 
 | | Crystal structure of fms1 in complex with S-N1-AcMeSpermidine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, N-{(1S)-3-[(4-aminobutyl)amino]-1-methylpropyl}acetamide, ... | | Authors: | Huang, Q, Hao, Q. | | Deposit date: | 2008-03-26 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of fms1 in complex with S-N1-AcMeSpermidine
TO BE PUBLISHED
|
|
3CND
 
 | | Crystal structure of fms1 in complex with N1-AcSpermine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[3-({4-[(3-aminopropyl)amino]butyl}amino)propyl]acetamide, Polyamine oxidase FMS1 | | Authors: | Huang, Q, Hao, Q. | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of fms1 in complex with N1-AcSpermine
TO BE PUBLISHED
|
|
3CNT
 
 | |
1MRH
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRK
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, ALPHA-TRICHOSANTHIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRG
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ADENOSINE, ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRI
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1MRJ
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ADENOSINE, ALPHA-TRICHOSANTHIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
1RSG
 
 | |
1EPT
 
 | | REFINED 1.8 ANGSTROMS RESOLUTION CRYSTAL STRUCTURE OF PORCINE EPSILON-TRYPSIN | | Descriptor: | CALCIUM ION, PORCINE E-TRYPSIN | | Authors: | Huang, Q, Wang, Z, Li, Y, Liu, S, Tang, Y. | | Deposit date: | 1994-06-07 | | Release date: | 1995-02-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined 1.8 A resolution crystal structure of the porcine epsilon-trypsin.
Biochim.Biophys.Acta, 1209, 1994
|
|
9E6X
 
 | | Crystal structure of ferritin | | Descriptor: | CADMIUM ION, FE (III) ION, Ferritin light chain, ... | | Authors: | Huang, Q. | | Deposit date: | 2024-10-31 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of ferritin
To Be Published
|
|
3CNS
 
 | |
1MG6
 
 | |
8DZT
 
 | | Crystal structure of human Sar1aH79G mutant | | Descriptor: | CALCIUM ION, GTP-binding protein SAR1a, GUANOSINE-5',3'-TETRAPHOSPHATE, ... | | Authors: | Huang, Q. | | Deposit date: | 2022-08-08 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The alarmone ppGpp selectively inhibits the isoform A of the human small GTPase Sar1.
Proteins, 91, 2023
|
|
8E0H
 
 | | Crystal structure of human Sar1aD104/D140A double mutant | | Descriptor: | GTP-binding protein SAR1a, GUANOSINE-5',3'-TETRAPHOSPHATE | | Authors: | Huang, Q. | | Deposit date: | 2022-08-09 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The alarmone ppGpp selectively inhibits the isoform A of the human small GTPase Sar1.
Proteins, 91, 2023
|
|
8E0A
 
 | | Crystal structure of human Sar1b | | Descriptor: | GTP-binding protein SAR1b, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Huang, Q. | | Deposit date: | 2022-08-08 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | The alarmone ppGpp selectively inhibits the isoform A of the human small GTPase Sar1.
Proteins, 91, 2023
|
|
8DZO
 
 | | Crystal structure of human Sar1T39N mutant | | Descriptor: | CALCIUM ION, GTP-binding protein SAR1a, GUANOSINE-5',3'-TETRAPHOSPHATE, ... | | Authors: | Huang, Q. | | Deposit date: | 2022-08-08 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The alarmone ppGpp selectively inhibits the isoform A of the human small GTPase Sar1.
Proteins, 91, 2023
|
|
8E0D
 
 | | Crystal structure of human Sar1bE140D | | Descriptor: | GTP-binding protein SAR1b, GUANOSINE-5',3'-TETRAPHOSPHATE | | Authors: | Huang, Q. | | Deposit date: | 2022-08-08 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | The alarmone ppGpp selectively inhibits the isoform A of the human small GTPase Sar1.
Proteins, 91, 2023
|
|
8E0B
 
 | | Crystal structure of human Sar1bT39N | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, Sar1bT39N | | Authors: | Huang, Q. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.212 Å) | | Cite: | The alarmone ppGpp selectively inhibits the isoform A of the human small GTPase Sar1.
Proteins, 91, 2023
|
|
8E0C
 
 | | Crystal structure of human Sar1bH79G | | Descriptor: | GTP-binding protein SAR1b, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Huang, Q. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | The alarmone ppGpp selectively inhibits the isoform A of the human small GTPase Sar1.
Proteins, 91, 2023
|
|
8DZN
 
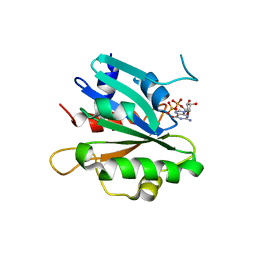 | | Crystal structure of human Sar1a in complex with GDP | | Descriptor: | GTP-binding protein SAR1a, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Huang, Q. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.107 Å) | | Cite: | The alarmone ppGpp selectively inhibits the isoform A of the human small GTPase Sar1.
Proteins, 91, 2023
|
|
8ETD
 
 | | Crystal Structure of Schizosaccharomyces pombe Rho1 | | Descriptor: | GTP-binding protein rho1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Huang, Q, Xie, J, Seetharaman, J. | | Deposit date: | 2022-10-16 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal Structure of Schizosaccharomyces pombe Rho1 Reveals Its Evolutionary Relationship with Other Rho GTPases.
Biology (Basel), 11, 2022
|
|
8DZM
 
 | |
1SSK
 
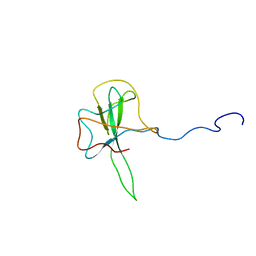 | | Structure of the N-terminal RNA-binding Domain of the SARS CoV Nucleocapsid Protein | | Descriptor: | Nucleocapsid protein | | Authors: | Huang, Q, Yu, L, Petros, A.M, Gunasekera, A, Liu, Z, Xu, N, Hajduk, P, Mack, J, Fesik, S.W, Olejniczak, E.T. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-Terminal RNA-Binding Domain of the SARS CoV Nucleocapsid Protein.
Biochemistry, 43, 2004
|
|
