7Q1T
 
 | | A de novo designed hetero-dimeric antiparallel coiled coil apCC-Di-AB | | Descriptor: | N-PROPANOL, SULFATE ION, apCC-Di-A, ... | | Authors: | Shanmugaratnam, S, Rhys, G.G, Dawson, W.M, Woolfson, D.N, Hocker, B. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | De novo designed peptides for cellular delivery and subcellular localisation.
Nat.Chem.Biol., 18, 2022
|
|
7Q1Q
 
 | | De novo designed homo-dimeric antiparallel helices Homomer-S | | Descriptor: | ACETATE ION, Homomer-S | | Authors: | Shanmugaratnam, S, Rhys, G.G, Dawson, W.M, Woolfson, D.N, Hocker, B. | | Deposit date: | 2021-10-20 | | Release date: | 2022-07-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | De novo designed peptides for cellular delivery and subcellular localisation.
Nat.Chem.Biol., 18, 2022
|
|
7PU4
 
 | |
7QSP
 
 | | Permutated C-terminal lobe of the ribose binding protein from Thermotoga maritima | | Descriptor: | 1,2-ETHANEDIOL, Ribose ABC transporter, periplasmic ribose-binding protein | | Authors: | Shanmugaratnam, S, Michel, F, Hocker, B. | | Deposit date: | 2022-01-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structures of permuted halves of a modern ribose-binding protein.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7QSQ
 
 | | Permutated N-terminal lobe of the ribose binding protein from Thermotoga maritima | | Descriptor: | 1,2-ETHANEDIOL, Ribose ABC transporter, periplasmic ribose-binding protein, ... | | Authors: | Shanmugaratnam, S, Michel, F, Hocker, B. | | Deposit date: | 2022-01-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structures of permuted halves of a modern ribose-binding protein.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
6YE0
 
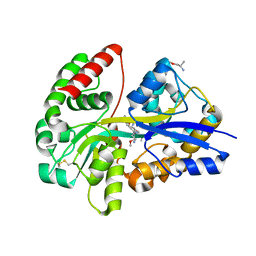 | | E.coli's Putrescine receptor PotF complexed with Putrescine | | Descriptor: | (2R)-1-methoxypropan-2-amine, (2~{R})-1-[(2~{R})-1-(2-methoxyethoxy)propan-2-yl]oxypropan-2-amine, (2~{S})-1-(2-methoxyethoxy)propan-2-amine, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2020-03-23 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF.
Structure, 29, 2021
|
|
6YEC
 
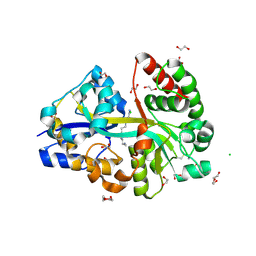 | | E.coli's Putrescine receptor PotF complexed with Spermine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2020-03-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF.
Structure, 29, 2021
|
|
6YQY
 
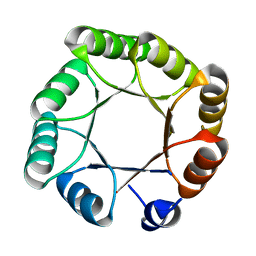 | | Crystal structure of sTIM11noCys, a de novo designed TIM barrel | | Descriptor: | de novo designed TIM barrel sTIM11noCys | | Authors: | Romero-Romero, S, Wiese, G.J, Kordes, S, Shanmugaratnam, S, Fernandez-Velasco, D.A, Hocker, B. | | Deposit date: | 2020-04-18 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | The Stability Landscape of de novo TIM Barrels Explored by a Modular Design Approach.
J.Mol.Biol., 433, 2021
|
|
6YQX
 
 | | Crystal structure of DeNovoTIM13, a de novo designed TIM barrel | | Descriptor: | CHLORIDE ION, GLYCEROL, de novo designed TIM barrel DeNovoTIM13 | | Authors: | Romero-Romero, S, Kordes, S, Shanmugaratnam, S, Fernandez-Velasco, D.A, Hocker, B. | | Deposit date: | 2020-04-18 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.638 Å) | | Cite: | The Stability Landscape of de novo TIM Barrels Explored by a Modular Design Approach.
J.Mol.Biol., 433, 2021
|
|
6Z2I
 
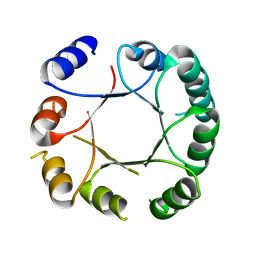 | | Crystal structure of DeNovoTIM6, a de novo designed TIM barrel | | Descriptor: | de novo designed TIM barrel DeNovoTIM6 | | Authors: | Romero-Romero, S, Kordes, S, Shanmugaratnam, S, Rodriguez-Romero, A, Fernandez-Velasco, D.A, Hocker, B. | | Deposit date: | 2020-05-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The Stability Landscape of de novo TIM Barrels Explored by a Modular Design Approach.
J.Mol.Biol., 433, 2021
|
|
7A8S
 
 | | de novo designed ba8-barrel sTIM11 with an alpha-helical extension | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, sTIM11_h3 | | Authors: | Shanmugaratnam, S, Wiese, J.G, Hocker, B. | | Deposit date: | 2020-08-31 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Extension of a de novo TIM barrel with a rationally designed secondary structure element.
Protein Sci., 30, 2021
|
|
6T1G
 
 | | Streptavidin variants harbouring an artificial organocatalyst based cofactor | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(1-pyridin-4-ylpiperidin-4-yl)pentanamide, Streptavidin | | Authors: | Lechner, H, Hocker, B. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Chembiochem, 22, 2021
|
|
6T1K
 
 | | Streptavidin variants harbouring an artificial organocatalyst based cofactor | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(1-pyridin-4-ylpiperidin-4-yl)pentanamide, Streptavidin | | Authors: | Lechner, H, Hocker, B. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Chembiochem, 22, 2021
|
|
6T2Z
 
 | | Streptavidin variants harbouring an artificial organocatalyst based cofactor | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(1-pyridin-4-ylpiperidin-4-yl)pentanamide, Streptavidin | | Authors: | Lechner, H, Hocker, B. | | Deposit date: | 2019-10-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Chembiochem, 22, 2021
|
|
6T2L
 
 | | Streptavidin variants harbouring an artificial organocatalyst based cofactor | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(1-pyridin-4-ylpiperidin-4-yl)pentanamide, GLYCEROL, ... | | Authors: | Lechner, H, Hocker, B. | | Deposit date: | 2019-10-09 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Chembiochem, 22, 2021
|
|
6T30
 
 | | Streptavidin variants harbouring an artificial organocatalyst based cofactor | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(1-pyridin-4-ylpiperidin-4-yl)pentanamide, GLYCEROL, ... | | Authors: | Lechner, H, Hocker, B. | | Deposit date: | 2019-10-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Chembiochem, 22, 2021
|
|
6TH8
 
 | |
6T32
 
 | | Streptavidin variants harbouring an artificial organocatalyst based cofactor | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(1-pyridin-4-ylpiperidin-4-yl)pentanamide, Streptavidin | | Authors: | Lechner, H, Hocker, B. | | Deposit date: | 2019-10-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Chembiochem, 22, 2021
|
|
6T31
 
 | | Streptavidin variants harbouring an artificial organocatalyst based cofactor | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(1-pyridin-4-ylpiperidin-4-yl)pentanamide, Streptavidin | | Authors: | Lechner, H, Hocker, B. | | Deposit date: | 2019-10-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Chembiochem, 22, 2021
|
|
6T2Y
 
 | | Streptavidin variants harbouring an artificial organocatalyst based cofactor | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(1-pyridin-4-ylpiperidin-4-yl)pentanamide, Streptavidin | | Authors: | Lechner, H, Hocker, B. | | Deposit date: | 2019-10-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Chembiochem, 22, 2021
|
|
7OYV
 
 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutations E39D F88A S247D in complex with spermidine | | Descriptor: | (2R)-1-methoxypropan-2-amine, (2~{R})-1-[(2~{R})-1-[(2~{R})-1-(2-methoxyethoxy)propan-2-yl]oxypropan-2-yl]oxypropan-2-amine, (2~{S})-1-methoxypropan-2-amine, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
7OYY
 
 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutation S247D in complex with spermidine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kroeger, P, Shanmugaratnam, S, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
7OYS
 
 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutations E39D Y87S in complex with spermidine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
7OYW
 
 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutations E39D F88L S247D in complex with spermidine | | Descriptor: | (2R)-1-methoxypropan-2-amine, (2~{R})-1-[(2~{R})-1-(2-methoxyethoxy)propan-2-yl]oxypropan-2-amine, (2~{S})-1-(2-methoxyethoxy)propan-2-amine, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
7OYT
 
 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutations E39D F88L in complex with spermidine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
