8U60
 
 | |
8U5K
 
 | |
1ZL3
 
 | | Coupling of active site motions and RNA binding | | Descriptor: | 5'-R(*GP*GP*CP*AP*AP*CP*GP*GP*UP*(FLO) UP*CP*GP*AP*UP*CP*CP*CP*GP*UP*UP*GP*C)-3', SULFATE ION, tRNA pseudouridine synthase B | | Authors: | Hoang, C, Hamilton, C.S, Mueller, E.G, Ferre-D'Amare, A.R. | | Deposit date: | 2005-05-05 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Precursor complex structure of pseudouridine synthase TruB suggests coupling of active site perturbations to an RNA-sequestering peripheral protein domain
Protein Sci., 14, 2005
|
|
2APO
 
 | | Crystal Structure of the Methanococcus jannaschii Cbf5 Nop10 Complex | | Descriptor: | POTASSIUM ION, Probable tRNA pseudouridine synthase B, Ribosome biogenesis protein Nop10, ... | | Authors: | Hamma, T, Reichow, S.L, Varani, G, Ferre-D'Amare, A.R. | | Deposit date: | 2005-08-16 | | Release date: | 2005-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Cbf5-Nop10 complex is a molecular bracket that organizes box H/ACA RNPs.
Nat.Struct.Mol.Biol., 12, 2005
|
|
3K1V
 
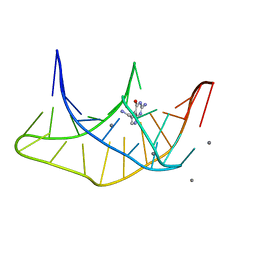 | |
3K0J
 
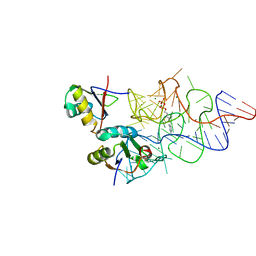 | | Crystal structure of the E. coli ThiM riboswitch in complex with thiamine pyrophosphate and the U1A crystallization module | | Descriptor: | MAGNESIUM ION, RNA (87-MER), THIAMINE DIPHOSPHATE, ... | | Authors: | Kulshina, N, Edwards, T.E, Ferre-D'Amare, A.R. | | Deposit date: | 2009-09-24 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch.
Rna, 16, 2010
|
|
3IWN
 
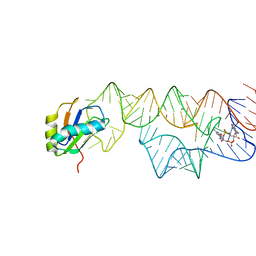 | | Co-crystal structure of a bacterial c-di-GMP riboswitch | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), C-di-GMP riboswitch, U1 small nuclear ribonucleoprotein A | | Authors: | Kulshina, N, Baird, N.J, Ferre-D'Amare, A.R. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2Z74
 
 | | T. tengcongensis glmS ribozyme bound to glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, glmS ribozyme RNA, ... | | Authors: | Klein, D.J, Wilkinson, S.R, Been, M.D, Ferre-D'Amare, A.R. | | Deposit date: | 2007-08-15 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Requirement of helix P2.2 and nucleotide G1 for positioning the cleavage site and cofactor of the glmS ribozyme
J.Mol.Biol., 373, 2007
|
|
3B4A
 
 | |
2Z75
 
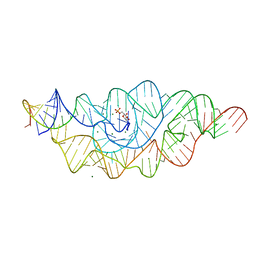 | | T. tengcongensis glmS ribozyme bound to glucosamine-6-phosphate | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, glmS ribozyme RNA, ... | | Authors: | Klein, D.J, Wilkinson, S.R, Been, M.D, Ferre-D'Amare, A.R. | | Deposit date: | 2007-08-15 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Requirement of helix P2.2 and nucleotide G1 for positioning the cleavage site and cofactor of the glmS ribozyme
J.Mol.Biol., 373, 2007
|
|
3B4C
 
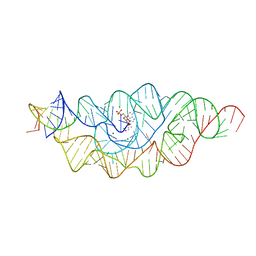 | |
3B4B
 
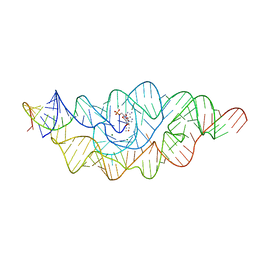 | |
3CUL
 
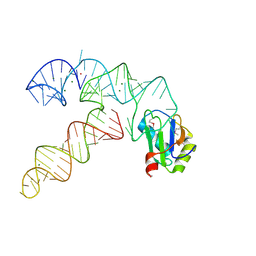 | | Aminoacyl-tRNA synthetase ribozyme | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (92-MER), ... | | Authors: | Xiao, H, Murakami, H, Suga, H, Ferre-D'Amare, A.R. | | Deposit date: | 2008-04-16 | | Release date: | 2008-06-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme.
Nature, 454, 2008
|
|
4TZW
 
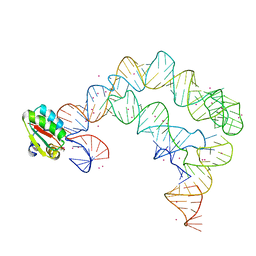 | | Co-crystals of the Ternary Complex Containing a T-box Stem I RNA, its Cognate tRNAGly, and B. subtilis YbxF protein, treated by removing lithium sulfate and replacing Mg2+ with Sr2+ post crystallization | | Descriptor: | Ribosome-associated protein L7Ae-like, STRONTIUM ION, T-box Stem I, ... | | Authors: | Zhang, J, Ferre-D'Amare, A.R. | | Deposit date: | 2014-07-11 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.671 Å) | | Cite: | Dramatic Improvement of Crystals of Large RNAs by Cation Replacement and Dehydration.
Structure, 22, 2014
|
|
8F5G
 
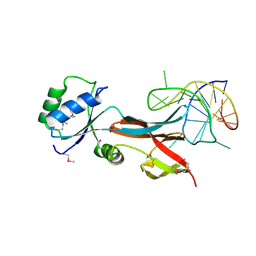 | | NusG-RNA complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, RNA, Transcription termination/antitermination protein NusG | | Authors: | Elghondakly, A.T, Ferre-D'Amare, A.R. | | Deposit date: | 2022-11-14 | | Release date: | 2023-11-22 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Major-groove sequence-specific RNA recognition by LoaP, a paralog of transcription elongation factor NusG.
Structure, 2024
|
|
8FI8
 
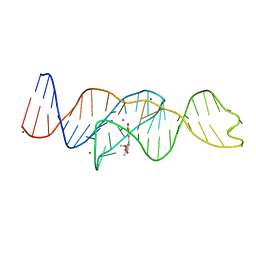 | |
8FHZ
 
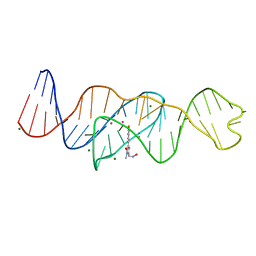 | | Structure of Lettuce aptamer bound to DFHO | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-12-15 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
8FHV
 
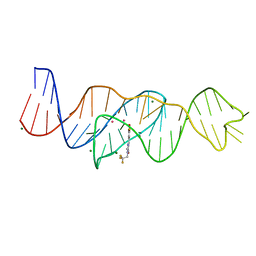 | | Structure of Lettuce aptamer bound to DFHBI-1T with thallium I ions | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-12-15 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
3CUN
 
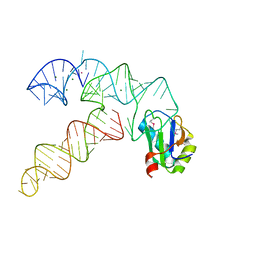 | | Aminoacyl-tRNA synthetase ribozyme | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Xiao, H, Murakami, H, Suga, H, Ferre-D'Amare, A.R. | | Deposit date: | 2008-04-16 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of specific tRNA aminoacylation by a small in vitro selected ribozyme.
Nature, 454, 2008
|
|
3FTM
 
 | |
8FI2
 
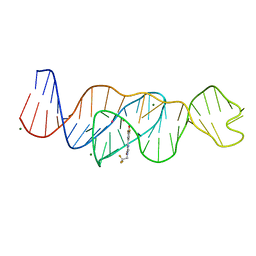 | | Structure of Lettuce C20T bound to DFHBI-1T | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-12-15 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
8FI1
 
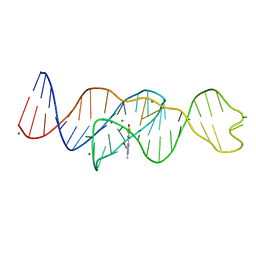 | | Structure of Lettuce C20G bound to DFHO | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-12-15 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
8FI0
 
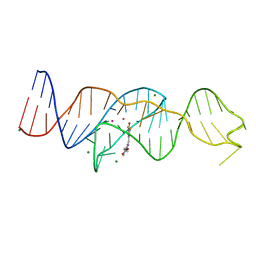 | |
8FI7
 
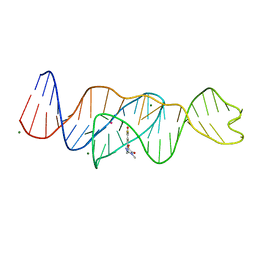 | | Structure of Lettuce C20T bound to DFHO | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-12-15 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
8FHX
 
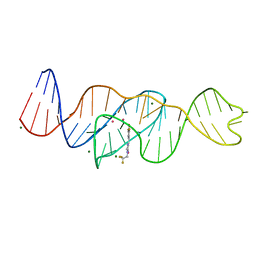 | | Structure of Lettuce aptamer bound to DFHBI-1T | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-12-15 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
