6SPP
 
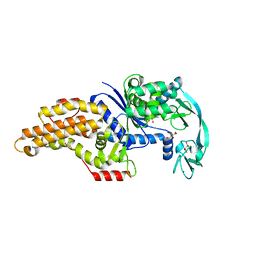 | | Structure of the Escherichia coli methionyl-tRNA synthetase variant VI298 | | Descriptor: | CITRIC ACID, GLYCEROL, Methionine--tRNA ligase, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
6SWD
 
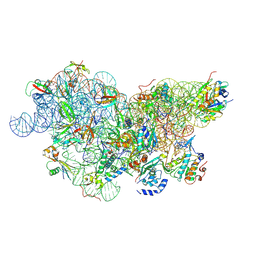 | | IC2 body model of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
6SPN
 
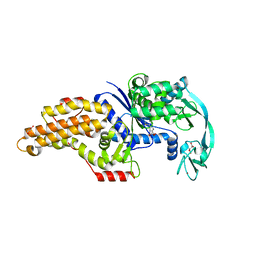 | | Structure of the Escherichia coli methionyl-tRNA synthetase complexed with beta-methionine | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
6SWC
 
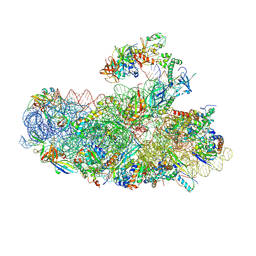 | | IC2B model of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
6SWE
 
 | | IC2 head of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S13, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
5OCD
 
 | |
6SPO
 
 | | Structure of the Escherichia coli methionyl-tRNA synthetase complexed with methionine | | Descriptor: | CITRIC ACID, GLYCEROL, METHIONINE, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
8BFW
 
 | | The structures of Ace2 in complex with bicyclic peptide inhibitor | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, ALA-CYS-VAL-ARG-SER-HIS-CYS-SER-SER-LEU-LEU-PRO-ARG-ILE-HIS-CYS-ALA-NH2, Processed angiotensin-converting enzyme 2 | | Authors: | Brear, P, Lulla, A, Harman, M, Dods, R, Chen, L, Bezerra, G, Demydchuk, Y, Stanway, S, Hyvonen, M. | | Deposit date: | 2022-10-27 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
J.Med.Chem., 66, 2023
|
|
6SPQ
 
 | | Structure of the Escherichia coli methionyl-tRNA synthetase variant VI298 complexed with methionine | | Descriptor: | CITRIC ACID, GLYCEROL, METHIONINE, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
6SW9
 
 | | IC2A model of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
8BN1
 
 | | The structures of Ace2 in complex with bicyclic peptide inhibitor | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, ALA-CYS-VAL-ARG-SER-4PH-CYS-SER-SER-LEU-LEU-PRO-ARG-ILE-HIS-CYS-ALA-NH2, Processed angiotensin-converting enzyme 2, ... | | Authors: | Brear, P, Lulla, A, Harman, M, Dods, R, Chen, L, Bezerra, G, Demydchuk, Y, Stanway, S, Hyvonen, M. | | Deposit date: | 2022-11-11 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
J.Med.Chem., 66, 2023
|
|
8B9P
 
 | | ACE2 in complex with bicyclic peptide inhibitor | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, ALA-CYS-GLY-ARG-GLN-PHE-CYS-HIS-THR-LEU-MET-PRO-ARG-HIS-LEU-CYS-ALA-NH2, Processed angiotensin-converting enzyme 2 | | Authors: | Brear, P, Lulla, A, Harman, M, Dods, R, Chen, L, Bezerra, G, Demydchuk, Y, Stanway, S, Hyvonen, M. | | Deposit date: | 2022-10-06 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
J.Med.Chem., 66, 2023
|
|
8BYJ
 
 | | The structures of Ace2 in complex with bicyclic peptide inhibitor | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, ALA-CYS-VAL-ARG-SER-HIS-CYS-SER-SER-LEU-LEU-PRO-ARG-ILE-HIS-CYS-ALA, Processed angiotensin-converting enzyme 2, ... | | Authors: | Brear, P, Lulla, A, Harman, M, Dods, R, Chen, L, Bezerra, G, Demydchuk, Y, Stanway, S, Hyvonen, M. | | Deposit date: | 2022-12-13 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
J.Med.Chem., 66, 2023
|
|
5L4O
 
 | |
6Y4B
 
 | |
6Y3G
 
 | | Crystal structure of phenylalanine tRNA from Escherichia coli | | Descriptor: | CALCIUM ION, GLYCEROL, GUANIDINE, ... | | Authors: | Bourgeois, G, Mechulam, Y, Schmitt, E. | | Deposit date: | 2020-02-18 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of the interaction between cyclodipeptide synthases and aminoacylated tRNA substrates.
Rna, 26, 2020
|
|
5MLQ
 
 | | Structure of CDPS from Nocardia brasiliensis | | Descriptor: | CDPS, CITRIC ACID | | Authors: | Bourgeois, G, Seguin, J, Moutiez, M, Babin, M, Belin, P, Mechulam, Y, Gondry, M, Schmitt, E. | | Deposit date: | 2016-12-07 | | Release date: | 2018-05-02 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structural basis for partition of the cyclodipeptide synthases into two subfamilies.
J.Struct.Biol., 203, 2018
|
|
5MLP
 
 | | Structure of CDPS from Rickettsiella grylli | | Descriptor: | Uncharacterized protein | | Authors: | Bourgeois, G, Seguin, J, Moutiez, M, Babin, M, Belin, P, Mechulam, Y, Gondry, M, Schmitt, E. | | Deposit date: | 2016-12-07 | | Release date: | 2018-05-02 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis for partition of the cyclodipeptide synthases into two subfamilies.
J.Struct.Biol., 203, 2018
|
|
6Q6R
 
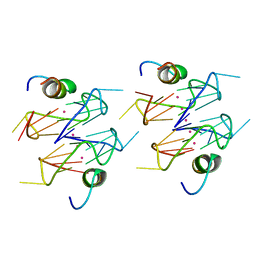 | | Recognition of different base tetrads by RHAU: X-ray crystal structure of G4 recognition motif bound to the 3-end tetrad of a DNA G-quadruplex | | Descriptor: | ATP-dependent DNA/RNA helicase DHX36, POTASSIUM ION, Parallel stranded DNA G-quadruplex | | Authors: | Heddi, B, Cheong, V.V, Schmitt, E, Mechulam, Y, Phan, A.T. | | Deposit date: | 2018-12-11 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of different base tetrads by RHAU (DHX36): X-ray crystal structure of the G4 recognition motif bound to the 3'-end tetrad of a DNA G-quadruplex.
J.Struct.Biol., 209, 2020
|
|
6QJO
 
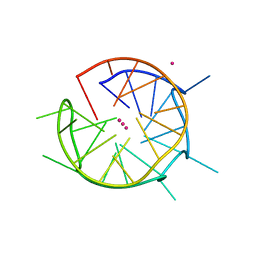 | | DNA containing both right- and left-handed parallel-stranded G-quadruplexes | | Descriptor: | DNA (28-MER), POTASSIUM ION | | Authors: | Winnerdy, F.R, Bakalar, B, Maity, A, Vandana, J.J, Schmitt, E, Mechulam, Y, Phan, A.T. | | Deposit date: | 2019-01-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NMR solution and X-ray crystal structures of a DNA molecule containing both right- and left-handed parallel-stranded G-quadruplexes.
Nucleic Acids Res., 47, 2019
|
|
5JB3
 
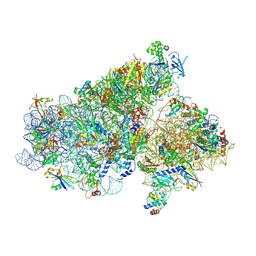 | | Cryo-EM structure of a full archaeal ribosomal translation initiation complex in the P-REMOTE conformation | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.-D, Schmitt, E, Mechulam, Y. | | Deposit date: | 2016-04-13 | | Release date: | 2016-11-30 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (5.34 Å) | | Cite: | Cryo-EM study of start codon selection during archaeal translation initiation.
Nat Commun, 7, 2016
|
|
5JBH
 
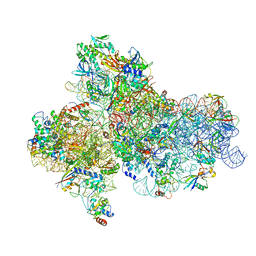 | | Cryo-EM structure of a full archaeal ribosomal translation initiation complex in the P-IN conformation | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein SX, 30S ribosomal protein eL41, ... | | Authors: | Coureux, P.-D, Schmitt, E, Mechulam, Y. | | Deposit date: | 2016-04-13 | | Release date: | 2016-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (5.34 Å) | | Cite: | Cryo-EM study of start codon selection during archaeal translation initiation.
Nat Commun, 7, 2016
|
|
4RCZ
 
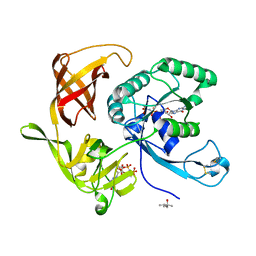 | | Structure of aIF2-gamma D19A variant from Sulfolobus solfataricus bound to GDPNP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Dubiez, E, Aleksandrov, A, Lazennec-Schurdevin, C, Mechulam, Y, Schmitt, E. | | Deposit date: | 2014-09-18 | | Release date: | 2015-05-27 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | Identification of a second GTP-bound magnesium ion in archaeal initiation factor 2.
Nucleic Acids Res., 43, 2015
|
|
4RD6
 
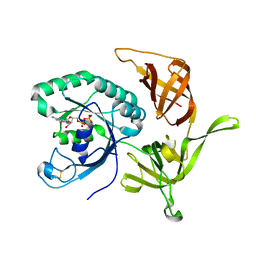 | | Structure of aIF2-gamma from Sulfolobus solfataricus bound to GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Translation initiation factor 2 subunit gamma | | Authors: | Dubiez, E, Aleksandrov, A, Lazennec-Schurdevin, C, Mechulam, Y, Schmitt, E. | | Deposit date: | 2014-09-18 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Identification of a second GTP-bound magnesium ion in archaeal initiation factor 2.
Nucleic Acids Res., 43, 2015
|
|
4RD1
 
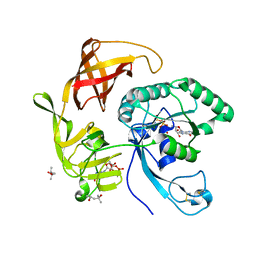 | | Structure of aIF2-gamma H97A variant from Sulfolobus solfataricus bound to GTP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Dubiez, E, Aleksandrov, A, Lazennec-Schurdevin, C, Mechulam, Y, Schmitt, E. | | Deposit date: | 2014-09-18 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of a second GTP-bound magnesium ion in archaeal initiation factor 2.
Nucleic Acids Res., 43, 2015
|
|
