1ETY
 
 | |
1ETW
 
 | |
4XRB
 
 | |
6J6T
 
 | | Crystal Structure of HDA15 HD domain | | Descriptor: | Histone deacetylase 15, POTASSIUM ION, SULFATE ION, ... | | Authors: | Cheng, Y.S, Hsu, J.C, Hung, H.C, Liu, T.C. | | Deposit date: | 2019-01-15 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure of Arabidopsis HISTONE DEACETYLASE15.
Plant Physiol., 184, 2020
|
|
5KVU
 
 | |
4XT8
 
 | | Crystal structure of Rv2671 from Mycobacterium tuberculosis in complex with NADP+ and trimetrexate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sacchettini, J.C, Cheng, Y.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-01-23 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into Mycobacterium tuberculosis Rv2671 Protein as a Dihydrofolate Reductase Functional Analogue Contributing to para-Aminosalicylic Acid Resistance.
Biochemistry, 55, 2016
|
|
4XT7
 
 | |
4XT6
 
 | | Crystal structure of Rv2671 from Mycobacterium tuberculosis in complex with the tetrahydropteridine ring of tetrahydrofolate (THF) | | Descriptor: | (6S)-2-amino-6-methyl-5,6,7,8-tetrahydropteridin-4(3H)-one, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Rv2671 | | Authors: | Sacchettini, J.C, Cheng, Y.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-01-23 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into Mycobacterium tuberculosis Rv2671 Protein as a Dihydrofolate Reductase Functional Analogue Contributing to para-Aminosalicylic Acid Resistance.
Biochemistry, 55, 2016
|
|
4XT5
 
 | |
4XT4
 
 | | Crystal structure of Rv2671 from Mycobacteirum tuberculosis in complex with dihydropteridine ring of dihydropteroic acid | | Descriptor: | 2-amino-6-methyl-7,8-dihydropteridin-4(3H)-one, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sacchettini, J.C, Cheng, Y.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-01-22 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Structural Insights into Mycobacterium tuberculosis Rv2671 Protein as a Dihydrofolate Reductase Functional Analogue Contributing to para-Aminosalicylic Acid Resistance.
Biochemistry, 55, 2016
|
|
5ECQ
 
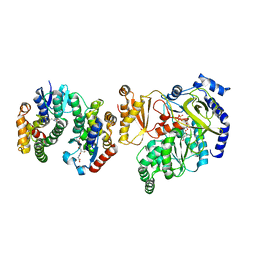 | | Crystal Structure of FIN219-FIP1 complex with JA, VAL and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTATHIONE, Glutathione S-transferase U20, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ECP
 
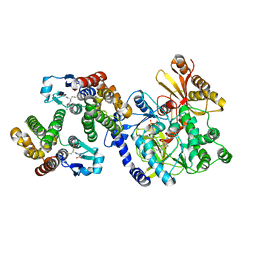 | | Crystal Structure of FIN219-FIP1 complex with JA, MET and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLUTATHIONE, Glutathione S-transferase U20, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25001574 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5ECS
 
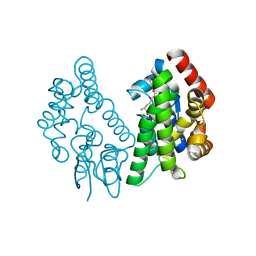 | | Crystal Structure of FIP1 with GSH | | Descriptor: | GLUTATHIONE, Glutathione S-transferase U20 | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6500026 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3AMS
 
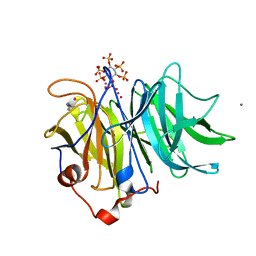 | | Crystal Structures of Bacillus subtilis Alkaline Phytase in Complex with Ca2+, Cd2+, Co2+, Ni2+, Mg2+ and myo-Inositol Hexasulfate | | Descriptor: | 3-phytase, CADMIUM ION, CALCIUM ION, ... | | Authors: | Zeng, Y.F, Ko, T.P, Lai, H.L, Cheng, Y.S, Wu, T.H, Ma, Y, Yang, C.S, Cheng, K.J, Huang, C.H, Guo, R.T, Liu, J.R. | | Deposit date: | 2010-08-22 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structures of Bacillus alkaline phytase in complex with divalent metal ions and inositol hexasulfate
J.Mol.Biol., 409, 2011
|
|
3AMR
 
 | | Crystal Structures of Bacillus subtilis Alkaline Phytase in Complex with Ca2+, Co2+, Ni2+, Mg2+ and myo-Inositol Hexasulfate | | Descriptor: | 3-phytase, CALCIUM ION, D-MYO-INOSITOL-HEXASULPHATE | | Authors: | Zeng, Y.F, Ko, T.P, Lai, H.L, Cheng, Y.S, Wu, T.H, Ma, Y, Yang, C.S, Cheng, K.J, Huang, C.H, Guo, R.T, Liu, J.R. | | Deposit date: | 2010-08-22 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of Bacillus alkaline phytase in complex with divalent metal ions and inositol hexasulfate
J.Mol.Biol., 409, 2011
|
|
3AZR
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellobiose | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AZS
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Mannotriose | | Descriptor: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AZT
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose | | Descriptor: | Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AXE
 
 | | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase V18Y/W203Y in complex with cellotetraose (cellobiose density was observed) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase, CALCIUM ION, ... | | Authors: | Huang, J.W, Cheng, Y.S, Ko, T.P, Lin, C.Y, Lai, H.L, Chen, C.C, Ma, Y, Huang, C.H, Zheng, Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-04-04 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Rational design to improve thermostability and specific activity of the truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase
Appl.Microbiol.Biotechnol., 94, 2012
|
|
3AXD
 
 | | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase V18Y/W203Y in apo-form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase, CALCIUM ION | | Authors: | Huang, J.W, Cheng, Y.S, Ko, T.P, Lin, C.Y, Lai, H.L, Chen, C.C, Ma, Y, Huang, C.H, Zheng, Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-04-03 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Rational design to improve thermostability and specific activity of the truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase
Appl.Microbiol.Biotechnol., 94, 2012
|
|
3WH9
 
 | | The ligand-free structure of ManBK from Aspergillus niger BK01 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huang, J.W, Chen, C.C, Huang, C.H, Huang, T.Y, Wu, T.H, Cheng, Y.S, Ko, T.P, Lin, C.Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2013-08-22 | | Release date: | 2014-10-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Analysis and Rational Design to Improve Specific Activity of beta-Mannanase from Aspergillus Niger BK01
To be Published
|
|
3WRE
 
 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
3WRG
 
 | | The complex structure of HypBA1 with L-arabinose | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase, ZINC ION, beta-L-arabinofuranose | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
3WRF
 
 | | The crystal structure of native HypBA1 from Bifidobacterium longum JCM 1217 | | Descriptor: | Non-reducing end beta-L-arabinofuranosidase | | Authors: | Huang, C.H, Zhu, Z, Cheng, Y.S, Chan, H.C, Ko, T.P, Chen, C.C, Wang, I, Ho, M.R, Hsu, S.T, Zeng, Y.F, Huang, Y.N, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-02-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and Catalytic Mechanism of a Glycoside Hydrolase Family-127 beta-L-Arabinofuranosidase (HypBA1)
J BIOPROCESS BIOTECH, 4, 2014
|
|
3WVJ
 
 | | The crystal structure of native glycosidic hydrolase | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase | | Authors: | Chen, C.C, Huang, J.W, Zhao, P, Ko, T.P, Huang, C.H, Chan, H.C, Huang, Z, Liu, W, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2014-05-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analyses and yeast production of the beta-1,3-1,4-glucanase catalytic module encoded by the licB gene of Clostridium thermocellum.
Enzyme.Microb.Technol., 71, 2015
|
|
