7XFT
 
 | | MucP PDZ2 domain | | Descriptor: | Putative zinc metalloprotease PA3649, SODIUM ION, VALINE | | Authors: | Lou, X, Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-04-02 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | MucP PDZ2 domain
To Be Published
|
|
7XFU
 
 | |
7XFS
 
 | | MucP PDZ1 domain | | Descriptor: | Putative zinc metalloprotease PA3649 | | Authors: | Lou, X, Li, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-04-02 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | MucP PDZ1 domain
To Be Published
|
|
5DV4
 
 | |
5YKR
 
 | |
5YKT
 
 | |
5Z2V
 
 | | Crystal structure of RecR from Pseudomonas aeruginosa PAO1 | | Descriptor: | ANY 5'-MONOPHOSPHATE NUCLEOTIDE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of RecR, a member of the RecFOR DNA-repair pathway, from Pseudomonas aeruginosa PAO1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5DV2
 
 | |
7X78
 
 | | L-fuculose 1-phosphate aldolase | | Descriptor: | L-fuculose phosphate aldolase, MAGNESIUM ION, SULFATE ION | | Authors: | Lou, X, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural characterization of an L-fuculose-1-phosphate aldolase from Klebsiella pneumoniae.
Biochem.Biophys.Res.Commun., 607, 2022
|
|
5WQN
 
 | |
5WQP
 
 | | Crystal structure of a carbonyl reductase from Pseudomonas aeruginosa PAO1 in complex with NADP (condition II) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE, PHOSPHATE ION, ... | | Authors: | Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2016-11-27 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and characterization of a NAD(P)H-dependent carbonyl reductase from Pseudomonas aeruginosa PAO1.
FEBS Lett., 591, 2017
|
|
5WQO
 
 | | Crystal structure of a carbonyl reductase from Pseudomonas aeruginosa PAO1 in complex with NADP (condition I) | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable dehydrogenase, ... | | Authors: | Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2016-11-27 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure and characterization of a NAD(P)H-dependent carbonyl reductase from Pseudomonas aeruginosa PAO1.
FEBS Lett., 591, 2017
|
|
5WQM
 
 | |
3OFU
 
 | | Crystal Structure of Cytochrome P450 CYP101C1 | | Descriptor: | (3E)-4-(2,6,6-trimethylcyclohex-1-en-1-yl)but-3-en-2-one, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhou, W, Ma, M, Bell, S.G, Yang, W, Hao, Y, Rees, N.H, Bartlam, M, Wong, L.-L, Rao, Z. | | Deposit date: | 2010-08-16 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444.
Chembiochem, 12, 2011
|
|
3OFT
 
 | | Crystal Structure of Cytochrome P450 CYP101C1 | | Descriptor: | (2R,5R)-hexane-2,5-diol, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhou, W, Ma, M, Bell, S.G, Yang, W, Hao, Y, Rees, N.H, Bartlam, M, Wong, L.-L, Rao, Z. | | Deposit date: | 2010-08-16 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444.
Chembiochem, 12, 2011
|
|
1NH9
 
 | | Crystal Structure of a DNA Binding Protein Mja10b from the hyperthermophile Methanococcus jannaschii | | Descriptor: | DNA-binding protein Alba | | Authors: | Wang, G, Bartlam, M, Guo, R, Yang, H, Xue, H, Liu, Y, Huang, L, Rao, Z. | | Deposit date: | 2002-12-19 | | Release date: | 2003-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a DNA binding protein from the hyperthermophilic euryarchaeon Methanococcus jannaschii
Protein Sci., 12, 2003
|
|
1P9E
 
 | | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3 | | Descriptor: | CADMIUM ION, Methyl Parathion Hydrolase, POTASSIUM ION, ... | | Authors: | Dong, Y, Sun, L, Bartlam, M, Rao, Z, Zhang, X. | | Deposit date: | 2003-05-11 | | Release date: | 2004-05-25 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of methyl parathion hydrolase from Pseudomonas sp. WBC-3.
J.Mol.Biol., 353, 2005
|
|
2Q6D
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease | | Descriptor: | Infectious bronchitis virus (IBV) main protease | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-04 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q6G
 
 | | Crystal structure of SARS-CoV main protease H41A mutant in complex with an N-terminal substrate | | Descriptor: | Polypeptide chain, severe acute respiratory syndrome coronavirus (SARS-CoV) | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2QC3
 
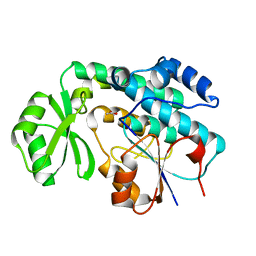 | | Crystal structure of MCAT from Mycobacterium tuberculosis | | Descriptor: | ACETIC ACID, Malonyl CoA-acyl carrier protein transacylase | | Authors: | Li, Z, Huang, Y, Ge, J, Bartlam, M, Wang, H, Rao, Z. | | Deposit date: | 2007-06-19 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of MCAT from Mycobacterium tuberculosis Reveals Three New Catalytic Models.
J.Mol.Biol., 371, 2007
|
|
2Q6F
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease in complex with a Michael acceptor inhibitor N3 | | Descriptor: | Infectious bronchitis virus (IBV) main protease, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
3NV5
 
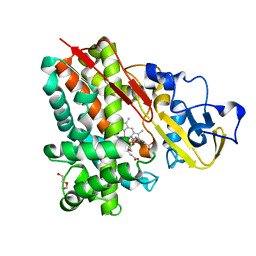 | | Crystal Structure of Cytochrome P450 CYP101D2 | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W.H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The structure of CYP101D2 unveils a potential path for substrate entry into the active site
Biochem.J., 433, 2011
|
|
4R0S
 
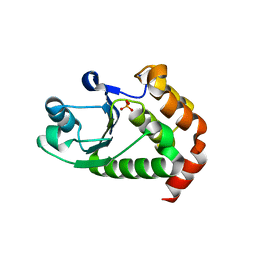 | | Crystal structure of P. aeruginosa TpbA | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein tyrosine phosphatase TpbA | | Authors: | Xu, K, Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural and Biochemical Analysis of Tyrosine Phosphatase Related to Biofilm Formation A (TpbA) from the Opportunistic Pathogen Pseudomonas aeruginosa PAO1
PLoS ONE, 10, 2015
|
|
3PSX
 
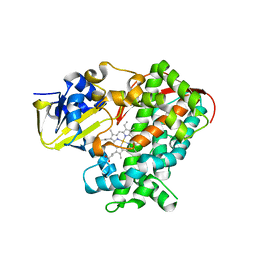 | | Crystal structure of the KT2 mutant of cytochrome P450 BM3 | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Whitehouse, C.J.C, Yorke, J.A, Bell, S.G, Zhou, W, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-12-02 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure, electronic properties and catalytic behaviour of an activity-enhancing CYP102A1 (P450(BM3)) variant
Dalton Trans, 40, 2011
|
|
4R0T
 
 | | Crystal structure of P. aeruginosa TpbA (C132S) in complex with pTyr | | Descriptor: | PHOSPHATE ION, Protein tyrosine phosphatase TpbA, TYROSINE | | Authors: | Xu, K, Li, S, Wang, Y, Bartlam, M. | | Deposit date: | 2014-08-01 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural and Biochemical Analysis of Tyrosine Phosphatase Related to Biofilm Formation A (TpbA) from the Opportunistic Pathogen Pseudomonas aeruginosa PAO1
Plos One, 10
|
|
