3J8B
 
 | | Model of the human eIF3 PCI-MPN octamer docked into the 43S-HCV IRES EM map | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit C, Eukaryotic translation initiation factor 3 subunit E, ... | | Authors: | Erzberger, J.P, Ban, N. | | Deposit date: | 2014-10-08 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell(Cambridge,Mass.), 158, 2014
|
|
3J8C
 
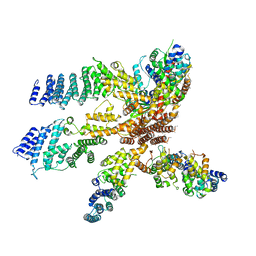 | | Model of the human eIF3 PCI-MPN octamer docked into the 43S EM map | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit C, Eukaryotic translation initiation factor 3 subunit E, ... | | Authors: | Erzberger, J.P, Ban, N. | | Deposit date: | 2014-10-08 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell(Cambridge,Mass.), 158, 2014
|
|
7P3A
 
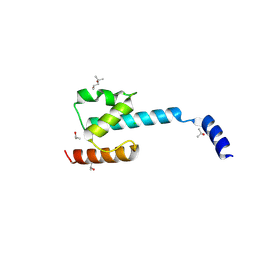 | | N-terminal domain of CGI-99 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Kroupova, A, Jinek, M. | | Deposit date: | 2021-07-07 | | Release date: | 2021-12-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular architecture of the human tRNA ligase complex.
Elife, 10, 2021
|
|
7P3B
 
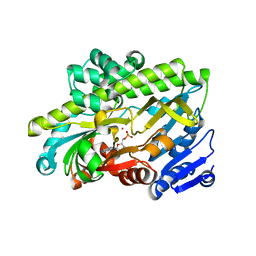 | | Human RNA ligase RTCB in complex with GMP and Co(II) | | Descriptor: | ACETATE ION, COBALT (II) ION, FORMIC ACID, ... | | Authors: | Kroupova, A, Ackle, F, Jinek, M. | | Deposit date: | 2021-07-07 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular architecture of the human tRNA ligase complex.
Elife, 10, 2021
|
|
4OUC
 
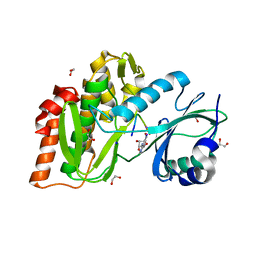 | | Structure of human haspin in complex with histone H3 substrate | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, Histone H3.2, ... | | Authors: | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-02-15 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of the chromatin phosphoproteome by the haspin protein kinase.
Mol Cell Proteomics, 13, 2014
|
|
5XBK
 
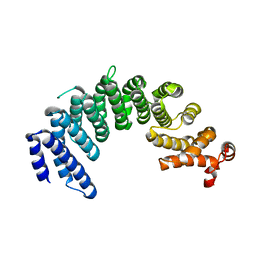 | | Crystal structure of human Importin4 | | Descriptor: | Importin-4, histone H3 | | Authors: | Song, J.J, Yoon, J. | | Deposit date: | 2017-03-20 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.223 Å) | | Cite: | Integrative Structural Investigation on the Architecture of Human Importin4_Histone H3/H4_Asf1a Complex and Its Histone H3 Tail Binding
J. Mol. Biol., 430, 2018
|
|
5XAH
 
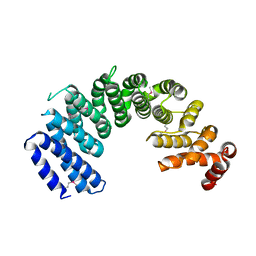 | | Crystal structure of human Importin4 | | Descriptor: | Importin-4 | | Authors: | Song, J.J, Yoon, J. | | Deposit date: | 2017-03-13 | | Release date: | 2018-02-14 | | Last modified: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Integrative Structural Investigation on the Architecture of Human Importin4_Histone H3/H4_Asf1a Complex and Its Histone H3 Tail Binding
J. Mol. Biol., 430, 2018
|
|
5NIN
 
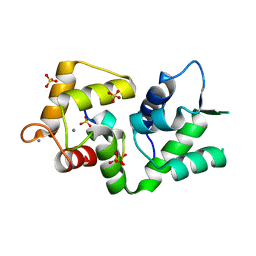 | |
4U1D
 
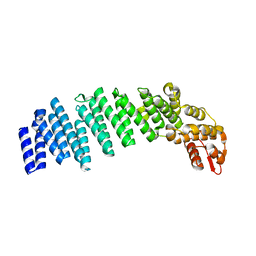 | |
4U1C
 
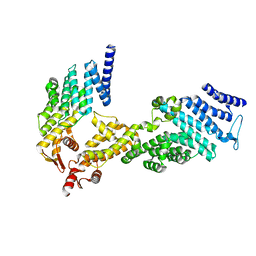 | |
4U1E
 
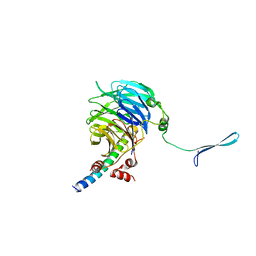 | | Crystal structure of the eIF3b-CTD/eIF3i/eIF3g-NTD translation initiation complex | | Descriptor: | Eukaryotic translation initiation factor 3 subunit B, Eukaryotic translation initiation factor 3 subunit G, Eukaryotic translation initiation factor 3 subunit I | | Authors: | Zhang, S, Erzberger, J.P, Schaefer, T, Ban, N. | | Deposit date: | 2014-07-15 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell, 158, 2014
|
|
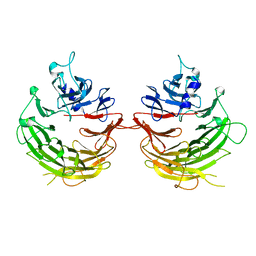
4U1F
 
 | |
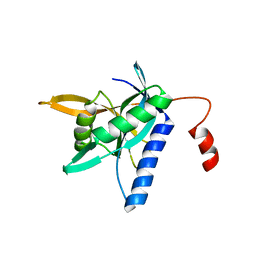
4QFT
 
 | |
6FUW
 
 | | Cryo-EM structure of the human CPSF160-WDR33-CPSF30 complex bound to the PAS AAUAAA motif at 3.1 Angstrom resolution | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, RNA (5'-R(P*AP*AP*UP*AP*AP*AP*GP*G)-3'), ... | | Authors: | Clerici, M, Faini, M, Jinek, M. | | Deposit date: | 2018-02-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of AAUAAA polyadenylation signal recognition by the human CPSF complex.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6G03
 
 | | NMR Solution Structure of yeast TSR2(1-152) | | Descriptor: | Pre-rRNA-processing protein TSR2 | | Authors: | Michel, E, Schuetz, S, Damberger, F.F, Allain, F.H.-T, Panse, V.G. | | Deposit date: | 2018-03-16 | | Release date: | 2018-09-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for disassembly of an importin:ribosomal protein complex by the escortin Tsr2.
Nat Commun, 9, 2018
|
|
6G04
 
 | | NMR Solution Structure of Yeast TSR2(1-152) in Complex with S26A(100-119) | | Descriptor: | 40S ribosomal protein S26-A, Pre-rRNA-processing protein TSR2 | | Authors: | Michel, E, Schuetz, S, Damberger, F.F, Allain, F.H.-T, Panse, V.G. | | Deposit date: | 2018-03-16 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for disassembly of an importin:ribosomal protein complex by the escortin Tsr2.
Nat Commun, 9, 2018
|
|
6F9N
 
 | | CRYSTAL STRUCTURE OF THE HUMAN CPSF160-WDR33 COMPLEX | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, pre-mRNA 3' end processing protein WDR33 | | Authors: | Clerici, M, Jinek, M. | | Deposit date: | 2017-12-14 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the assembly and polyA signal recognition mechanism of the human CPSF complex.
Elife, 6, 2017
|
|
6FEC
 
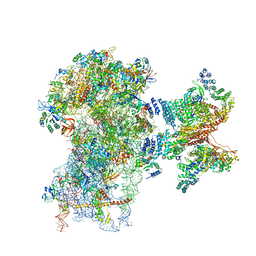 | | Human cap-dependent 48S pre-initiation complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schaffitzel, C, Schaffitzel, C. | | Deposit date: | 2017-12-31 | | Release date: | 2018-03-14 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structure of a human cap-dependent 48S translation pre-initiation complex.
Nucleic Acids Res., 46, 2018
|
|
6FBL
 
 | | NMR Solution Structure of MINA-1(254-334) | | Descriptor: | MINA-1 | | Authors: | Michel, E, Allain, F. | | Deposit date: | 2017-12-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | MINA-1 and WAGO-4 are part of regulatory network coordinating germ cell death and RNAi in C. elegans.
Cell Death Differ., 26, 2019
|
|
6EMK
 
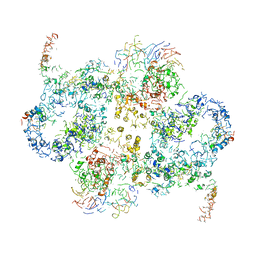 | | Cryo-EM Structure of Saccharomyces cerevisiae Target of Rapamycin Complex 2 | | Descriptor: | Serine/threonine-protein kinase TOR2, Target of rapamycin complex 2 subunit AVO1, Target of rapamycin complex 2 subunit AVO2, ... | | Authors: | Karuppasamy, M, Kusmider, B, Oliveira, T.M, Gaubitz, C, Prouteau, M, Loewith, R, Schaffitzel, C. | | Deposit date: | 2017-10-02 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Cryo-EM structure of Saccharomyces cerevisiae target of rapamycin complex 2.
Nat Commun, 8, 2017
|
|
6OIS
 
 | | CryoEM structure of Arabidopsis DR complex (DMS3-RDM1) | | Descriptor: | Protein DEFECTIVE IN MERISTEM SILENCING 3, Protein RDM1 | | Authors: | Wongpalee, S.P, Liu, S, Zhou, Z.H, Jacobsen, S.E. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CryoEM structures of Arabidopsis DDR complexes involved in RNA-directed DNA methylation.
Nat Commun, 10, 2019
|
|
6OIT
 
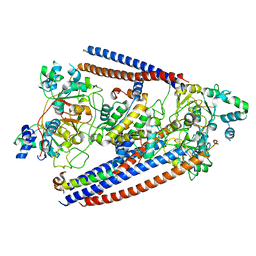 | | CryoEM structure of Arabidopsis DDR' complex (DRD1 peptide-DMS3-RDM1) | | Descriptor: | Protein CHROMATIN REMODELING 35, Protein DEFECTIVE IN MERISTEM SILENCING 3, Protein RDM1 | | Authors: | Wongpalee, S.P, Liu, S, Zhou, Z.H, Jacobsen, S.E. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | CryoEM structures of Arabidopsis DDR complexes involved in RNA-directed DNA methylation.
Nat Commun, 10, 2019
|
|
