7OAU
 
 | | Nanobody C5 bound to Kent variant RBD (N501Y) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C5, GLYCEROL, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7QUR
 
 | | SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry | | Descriptor: | 2,3,5-tris(iodanyl)benzamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
7QUS
 
 | | SARS-CoV-2 Spike, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
1DZT
 
 | | RMLC FROM SALMONELLA TYPHIMURIUM | | Descriptor: | 3'-O-ACETYLTHYMIDINE-(5' DIPHOSPHATE PHENYL ESTER), 3'-O-ACETYLTHYMIDINE-5'-DIPHOSPHATE, DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, ... | | Authors: | Naismith, J.H, Giraud, M.F. | | Deposit date: | 2000-03-07 | | Release date: | 2000-04-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rmlc, the Third Enzyme of Dtdp-L-Rhamnose Pathway, is a New Class of Epimerase.
Nat.Struct.Biol., 7, 2000
|
|
1DZR
 
 | | RmlC from Salmonella typhimurium | | Descriptor: | DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, GLYCEROL, SULFATE ION | | Authors: | Naismith, J.H, Giraud, M.F. | | Deposit date: | 2000-03-07 | | Release date: | 2000-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Rmlc, the Third Enzyme of Dtdp-L-Rhamnose Pathway, is a New Class of Epimerase.
Nat.Struct.Biol., 7, 2000
|
|
5N0O
 
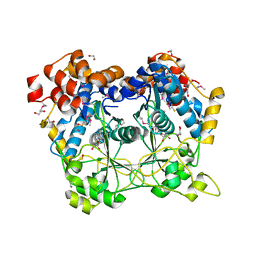 | |
6ENK
 
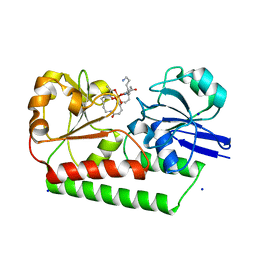 | | The X-ray crystal structure of DesE bound to desferrioxamine B | | Descriptor: | DesE, SODIUM ION, desferrioxamine B | | Authors: | Naismith, J.H, McMahon, S.A, Challis, G.L, Kadi, N, Oke, M, Liu, H, Carter, L.G, Johnson, K.A. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Desferrioxamine biosynthesis: diverse hydroxamate assembly by substrate-tolerant acyl transferase DesC.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
6FI2
 
 | | VexL: A periplasmic depolymerase provides new insight into ABC transporter-dependent secretion of bacterial capsular polysaccharides | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranuronic acid-(1-4)-3-O-acetyl-2-acetamido-2-deoxy-alpha-D-galactopyranuronic acid-(1-4)-3-O-acetyl-2-acetamido-2-deoxy-alpha-D-galactopyranuronic acid, MALONATE ION, VexL | | Authors: | Naismith, J.H, McMahon, S.A, Le Bas, A, Liston, S.D, Whitfield, C. | | Deposit date: | 2018-01-16 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Periplasmic depolymerase provides insight into ABC transporter-dependent secretion of bacterial capsular polysaccharides.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5N0P
 
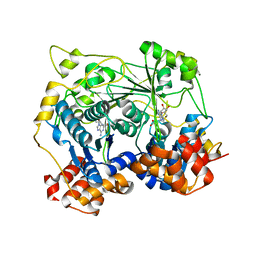 | | Crystal structure of OphA-DeltaC18 in complex with SAH | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, peptide N-methyltranferase | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A molecular mechanism for the enzymatic methylation of nitrogen atoms within peptide bonds.
Sci Adv, 4, 2018
|
|
5N0Q
 
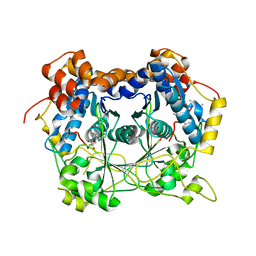 | |
6Z3C
 
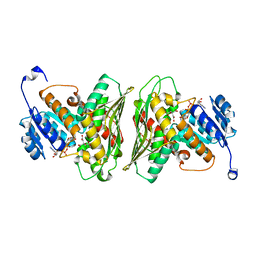 | | High resolution structure of RgNanOx | | Descriptor: | CITRATE ANION, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Naismith, J.H, Lee, M.O. | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Uncovering a novel molecular mechanism for scavenging sialic acids in bacteria.
J.Biol.Chem., 295, 2020
|
|
6Z33
 
 | |
6Z3B
 
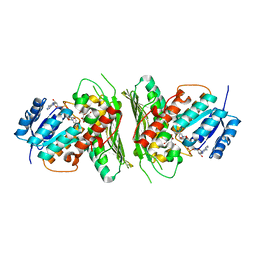 | | Low resolution structure of RgNanOx | | Descriptor: | CITRIC ACID, Gfo/Idh/MocA family oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Naismith, J.H, Lee, M. | | Deposit date: | 2020-05-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Uncovering a novel molecular mechanism for scavenging sialic acids in bacteria.
J.Biol.Chem., 295, 2020
|
|
6Z2N
 
 | |
6ZHD
 
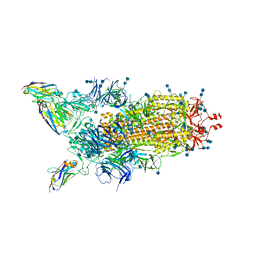 | | H11-H4 bound to Spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody H11-H4, ... | | Authors: | Clare, D.K, Naismith, J.H, Weckener, M, Vogirala, V.K. | | Deposit date: | 2020-06-22 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | H11-H4 bound to Spike
To Be Published
|
|
6YY5
 
 | | Crystal structure of the ferric enterobactin receptor (PfeA) in complex with TCV_L5 | | Descriptor: | FE (III) ION, Ferric enterobactin receptor, ~{N}-[2-[[(2~{S})-3-[[(2~{S})-3-[[1-[2-[2-[2-[4-[4-[5-(acetamidomethyl)-2-oxidanylidene-1,3-oxazolidin-3-yl]-2-fluoranyl-phenyl]piperazin-1-yl]-2-oxidanylidene-ethoxy]ethoxy]ethyl]-1,2,3-triazol-4-yl]methylamino]-2-[[2,3-bis(oxidanyl)phenyl]carbonylamino]-3-oxidanylidene-propyl]amino]-2-[[2,3-bis(oxidanyl)phenyl]carbonylamino]-3-oxidanylidene-propyl]amino]-2-oxidanylidene-ethyl]-2,3-bis(oxidanyl)benzamide | | Authors: | Naismith, J.H, Moynie, L.M. | | Deposit date: | 2020-05-04 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | Hijacking of the Enterobactin Pathway by a Synthetic Catechol Vector Designed for Oxazolidinone Antibiotic Delivery in Pseudomonas aeruginosa.
Acs Infect Dis., 2022
|
|
6ZH9
 
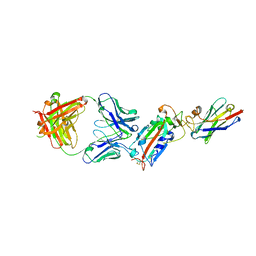 | | Ternary complex CR3022 H11-H4 and RBD (SARS-CoV-2) | | Descriptor: | CR3022 Light chain, CR3022 heavy, Nanobody H11-H4, ... | | Authors: | Naismith, J.H, Mikolajek, H, Le Bas, A. | | Deposit date: | 2020-06-21 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6YQQ
 
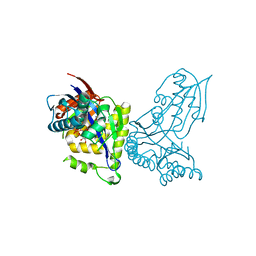 | | ForT-PRPP complex | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, CHLORIDE ION, ForT-PRPP complex, ... | | Authors: | Naismith, J.H, Gao, S. | | Deposit date: | 2020-04-18 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Uncovering the chemistry of C-C bond formation in C-nucleoside biosynthesis: crystal structure of a C-glycoside synthase/PRPP complex.
Chem.Commun.(Camb.), 56, 2020
|
|
6Y47
 
 | |
1TEI
 
 | | STRUCTURE OF CONCANAVALIN A COMPLEXED TO BETA-D-GLCNAC (1,2)ALPHA-D-MAN-(1,6)[BETA-D-GLCNAC(1,2)ALPHA-D-MAN (1,6)]ALPHA-D-MAN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, CALCIUM ION, CONCANAVALIN A, ... | | Authors: | Naismith, J.H, Moothoo, D.N. | | Deposit date: | 1997-05-28 | | Release date: | 1997-08-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Concanavalin A distorts the beta-GlcNAc-(1-->2)-Man linkage of beta-GlcNAc-(1-->2)-alpha-Man-(1-->3)-[beta-GlcNAc-(1-->2)-alpha-Man- (1-->6)]-Man upon binding.
Glycobiology, 8, 1998
|
|
1A3S
 
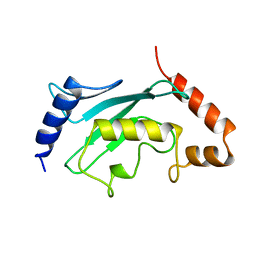 | | HUMAN UBC9 | | Descriptor: | UBC9 | | Authors: | Naismith, J.H, Giraud, M. | | Deposit date: | 1998-01-23 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of ubiquitin-conjugating enzyme 9 displays significant differences with other ubiquitin-conjugating enzymes which may reflect its specificity for sumo rather than ubiquitin.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
5H8C
 
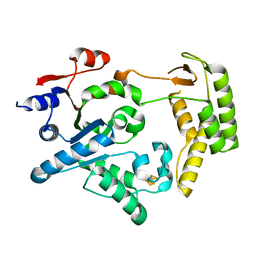 | | Truncated XPD | | Descriptor: | IRON/SULFUR CLUSTER, XPD/Rad3 related DNA helicase | | Authors: | Naismith, J.H, Constantinescu, D. | | Deposit date: | 2015-12-23 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Mechanism of DNA loading by the DNA repair helicase XPD.
Nucleic Acids Res., 44, 2016
|
|
5H8W
 
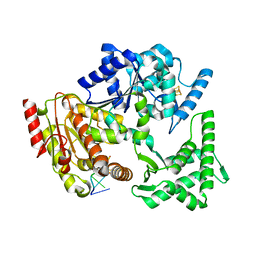 | | XPD mechanism | | Descriptor: | ATP-dependent DNA helicase Ta0057, DNA (5'-D(P*TP*AP*CP*GP*A)-3'), IRON/SULFUR CLUSTER, ... | | Authors: | Naismith, J.H, Constantinescu, D. | | Deposit date: | 2015-12-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of DNA loading by the DNA repair helicase XPD.
Nucleic Acids Res., 44, 2016
|
|
6SJ4
 
 | | Amidohydrolase, AHS with substrate analog | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-hydroxyphenyl)carbonyloxybenzoic acid, Amidohydrolase, ... | | Authors: | Naismith, J.H, Song, H. | | Deposit date: | 2019-08-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The Biosynthesis of the Benzoxazole in Nataxazole Proceeds via an Unstable Ester and has Synthetic Utility.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5AJI
 
 | | MscS D67R1 high resolution | | Descriptor: | DECANE, HEXANE, N-OCTANE, ... | | Authors: | Naismith, J.H, Pliotas, C. | | Deposit date: | 2015-02-24 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Role of Lipids in Mechanosensation.
Nat.Struct.Mol.Biol., 22, 2015
|
|
