8BHW
 
 | |
6RBG
 
 | |
6NWR
 
 | |
7QBP
 
 | | Crystal structure of R2-like ligand-binding oxidase from Saccharopolyspora Erythraea | | Descriptor: | FE (III) ION, MANGANESE (III) ION, PALMITIC ACID, ... | | Authors: | Srinivas, V, Diamanti, R, Lebrette, H, Hogbom, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Comparative structural analysis provides new insights into the function of R2-like ligand-binding oxidase.
Febs Lett., 596, 2022
|
|
8P3O
 
 | |
8P3P
 
 | |
7BET
 
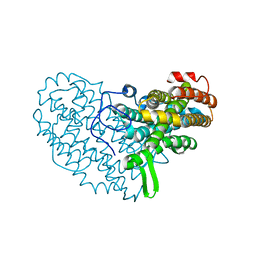 | | Structure of Ribonucleotide reductase R2 from Escherichia coli collected by femtosecond serial crystallography on a COC membrane | | Descriptor: | FE (III) ION, Ribonucleoside-diphosphate reductase 1 subunit beta | | Authors: | Aurelius, O, John, J, Martiel, I, Marsh, M, Vera, L, Huang, C.Y, Olieric, V, Leonarski, P, Nass, K, Padeste, C, Karpik, A, Hogbom, M, Wang, M, Pedrini, B. | | Deposit date: | 2020-12-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Commissioning results from the SwissMX instrument for fixed target macromolecular crystallography at SwissFEL
To Be Published
|
|
1RSV
 
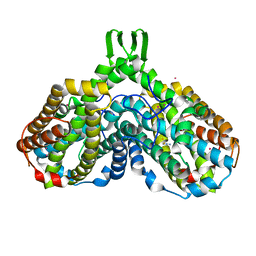 | | azide complex of the diferrous E238A mutant R2 subunit of ribonucleotide reductase | | Descriptor: | AZIDE ION, FE (III) ION, MERCURY (II) ION, ... | | Authors: | Assarsson, M, Andersson, M.E, Hogbom, M, Persson, B.O, Sahlin, M, Barra, A.L, Sjoberg, B.M, Nordlund, P, Graslund, A. | | Deposit date: | 2003-12-10 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Restoring proper radical generation by azide binding to the iron site of the E238A mutant R2 protein of ribonucleotide reductase from Escherichia coli.
J.Biol.Chem., 276, 2001
|
|
7BG4
 
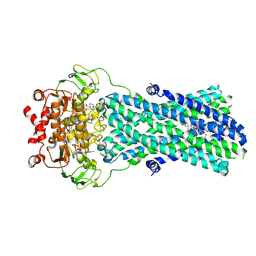 | | Multidrug resistance transporter BmrA mutant E504A bound with ATP, Mg, and Rhodamine 6G solved by Cryo-EM | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid A export ATP-binding/permease protein MsbA, MAGNESIUM ION, ... | | Authors: | Wiseman, B, Chaptal, V, Zampieri, V, Magnard, S, Hogbom, M, Falson, P. | | Deposit date: | 2021-01-05 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
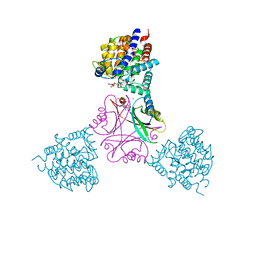
5OVO
 
 | |
5OMK
 
 | |
3GZD
 
 | | Human selenocysteine lyase, P1 crystal form | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, NITRATE ION, Selenocysteine lyase | | Authors: | Karlberg, T, Hogbom, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-07 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical discrimination between selenium and sulfur 1: a single residue provides selenium specificity to human selenocysteine lyase.
Plos One, 7, 2012
|
|
3GZC
 
 | | Structure of human selenocysteine lyase | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, Selenocysteine lyase | | Authors: | Collins, R, Hogbom, M, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Karlberg, T, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Uppenberg, J, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-07 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical discrimination between selenium and sulfur 1: a single residue provides selenium specificity to human selenocysteine lyase.
Plos One, 7, 2012
|
|
3R44
 
 | | Mycobacterium tuberculosis fatty acyl CoA synthetase | | Descriptor: | HISTIDINE, MALONATE ION, fatty acyl CoA synthetase FADD13 (FATTY-ACYL-CoA SYNTHETASE) | | Authors: | Andersson, C.S, Martinez Molina, D, Hogbom, M. | | Deposit date: | 2011-03-17 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Mycobacterium tuberculosis Very-Long-Chain Fatty Acyl-CoA Synthetase: Structural Basis for Housing Lipid Substrates Longer than the Enzyme.
Structure, 20, 2012
|
|
2UW2
 
 | | Crystal structure of human ribonucleotide reductase subunit R2 | | Descriptor: | FE (III) ION, RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE M2 SUBUNIT | | Authors: | Welin, M, Ogg, D, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Kotenyova, T, Magnusdottir, A, Moche, M, Nilsson-Ehle, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Stenmark, P, Uppenberg, J, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Nordlund, P. | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Ribonucleotide Reductase Subunit R2
To be Published
|
|
2UVQ
 
 | | Crystal structure of human uridine-cytidine kinase 1 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, URIDINE-CYTIDINE KINASE 1 | | Authors: | Kosinska, U, Stenmark, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E.P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Uppsten, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2007-03-13 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Human Uridine-Cytidine Kinase 1
To be Published
|
|
7QBK
 
 | | Crystal structure of a second homolog of R2-like ligand-binding oxidase in Sulfolobus acidocaldarius (SaR2loxII) | | Descriptor: | FE (III) ION, MANGANESE (III) ION, R2-like ligand-binding oxidase (homolog II) from Sulfolobus acidocaldarius | | Authors: | Lebrette, H, Diamanti, R, Srinivas, V, Hogbom, M. | | Deposit date: | 2021-11-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Comparative structural analysis provides new insights into the function of R2-like ligand-binding oxidase.
Febs Lett., 596, 2022
|
|
6Y2N
 
 | | Crystal structure of ribonucleotide reductase R2 subunit solved by serial synchrotron crystallography | | Descriptor: | FE (III) ION, MANGANESE (III) ION, Ribonucleoside-diphosphate reductase subunit beta | | Authors: | Shilova, A, Lebrette, H, Aurelius, O, Hogbom, M, Mueller, U. | | Deposit date: | 2020-02-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Current status and future opportunities for serial crystallography at MAX IV Laboratory.
J.Synchrotron Radiat., 27, 2020
|
|
2A5V
 
 | | Crystal structure of M. tuberculosis beta carbonic anhydrase, Rv3588c, tetrameric form | | Descriptor: | CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE), THIOCYANATE ION, ZINC ION | | Authors: | Covarrubias, A.S, Bergfors, T, Jones, T.A, Hogbom, M. | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Mechanics of the pH-dependent Activity of beta-Carbonic Anhydrase from Mycobacterium tuberculosis
J.Biol.Chem., 281, 2006
|
|
1RSR
 
 | | azide complex of the diferrous F208A mutant R2 subunit of ribonucleotide reductase | | Descriptor: | AZIDE ION, FE (II) ION, MERCURY (II) ION, ... | | Authors: | Andersson, M.E, Hogbom, M, Rinaldo-Matthis, A, Andersson, K.K, Sjoberg, B.M, Nordlund, P. | | Deposit date: | 2003-12-10 | | Release date: | 2003-12-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of an Azide Complex of the Diferrous R2 Subunit of Ribonucleotide Reductase Displays a Novel Carboxylate Shift with Important Mechanistic Implications for Diiron-Catalyzed Oxygen Activation
J.Am.Chem.Soc., 121, 1999
|
|
4HR4
 
 | |
4HR5
 
 | | R2-like ligand-binding oxidase without metal cofactor | | Descriptor: | PALMITIC ACID, Ribonuleotide reductase small subunit | | Authors: | Griese, J.J, Hogbom, M. | | Deposit date: | 2012-10-26 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | Direct observation of structurally encoded metal discrimination and ether bond formation in a heterodinuclear metalloprotein
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HR0
 
 | |
7AI9
 
 | | Structure of Ribonucleotide reductase R2 from Escherichia coli collected by rotation serial crystallography on a COC membrane at a synchrotron source | | Descriptor: | FE (III) ION, Ribonucleoside-diphosphate reductase 1 subunit beta | | Authors: | Aurelius, O, John, J, Martiel, I, Padeste, C, Karpik, A, Huang, C.Y, Hogbom, M, Wang, M, Marsh, M. | | Deposit date: | 2020-09-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Versatile microporous polymer-based supports for serial macromolecular crystallography.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7AI8
 
 | | Structure of Ribonucleotide reductase R2 from Escherichia coli collected by still serial crystallography on a COC membrane at a synchrotron source | | Descriptor: | FE (III) ION, Ribonucleoside-diphosphate reductase 1 subunit beta | | Authors: | Aurelius, O, John, J, Martiel, I, Padeste, C, Karpik, A, Huang, C.Y, Hogbom, M, Wang, M, Marsh, M. | | Deposit date: | 2020-09-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Versatile microporous polymer-based supports for serial macromolecular crystallography.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
