7R6S
 
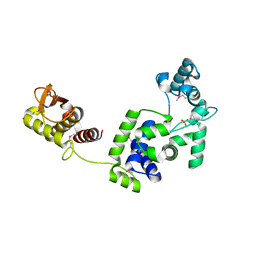 | | Crystal Structure of the Putative Bacteriophage Protein from Stenotrophomonas maltophilia | | Descriptor: | Putative bacteriophage protein, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Brunzelle, J.S, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2021-06-23 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Putative Bacteriophage Protein from Stenotrophomonas maltophilia
To Be Published
|
|
8G1Y
 
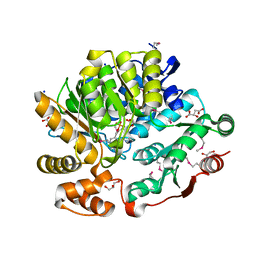 | | Crystal Structure of the Threonine Synthase from Streptococcus pneumoniae in complex with Pyridoxal 5-phosphate. | | Descriptor: | CHLORIDE ION, D-MALATE, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-02-03 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal Structure of the Threonine Synthase from Streptococcus pneumoniae in complex with Pyridoxal 5-phosphate.
To Be Published
|
|
8G28
 
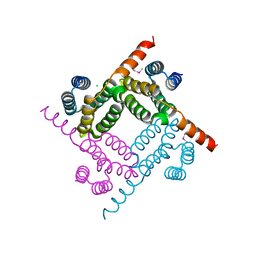 | | Crystal Structure of the C-terminal Fragment of AAA ATPase from Streptococcus pneumoniae. | | Descriptor: | ATPase, AAA family, CHLORIDE ION | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of the C-terminal Fragment of AAA ATPase from Streptococcus pneumoniae.
To Be Published
|
|
8G22
 
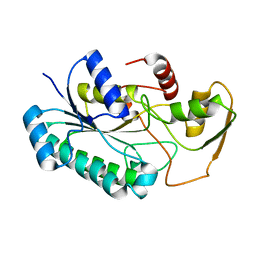 | | Crystal Structure of the dTDP-4-dehydrorhamnose Reductase from Streptococcus pneumoniae. | | Descriptor: | dTDP-4-dehydrorhamnose reductase | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-02-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structure of the dTDP-4-dehydrorhamnose Reductase from Streptococcus pneumoniae.
To Be Published
|
|
8G62
 
 | | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004 | | Descriptor: | 3-methoxy-5-(1-methylpiperidin-4-yl)-N-[4-(pyrrolidine-1-sulfonyl)phenyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Luci, D, Kales, S, Simeonov, A, Rai, G, Drayman, N, Tay, S, Oakes, S, Rosner, M, Chen, B, Dulin, N, Solway, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-02-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004
To Be Published
|
|
8EY4
 
 | | Contact-dependent growth inhibition toxin-immunity protein complex from E. coli O32:H37 | | Descriptor: | Cys_rich_CPCC domain-containing protein, FE (III) ION, PT-VENN domain-containing protein | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Hayes, C.S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Contact-dependent growth inhibition toxin-immunity protein complex from E. coli O32:H37
To Be Published
|
|
3E7N
 
 | | Crystal structure of d-ribose high-affinity transport system from salmonella typhimurium lt2 | | Descriptor: | 1,2-ETHANEDIOL, D-ribose high-affinity transport system | | Authors: | Nocek, B, Maltseva, N, Gu, M, Joachimiak, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-18 | | Release date: | 2008-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of d-ribose high-affinity transport system from salmonella typhimurium lt2
To be Published
|
|
8F8O
 
 | | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic and L-Lactic Acids | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, CITRIC ACID, SUCCINIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Pshenychnyi, S, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-11-22 | | Release date: | 2022-11-30 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Succinyl-diaminopimelate Desuccinylase (DapE) from Acinetobacter baumannii in complex with Succinic and L-Lactic Acids
To Be Published
|
|
8EBC
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria monocytogenes in the complex with IMP | | Descriptor: | FORMIC ACID, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Osipiuk, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-08-31 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria monocytogenes in the complex with IMP
To Be Published
|
|
6OZ7
 
 | | Putative oxidoreductase from Escherichia coli str. K-12 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Osipiuk, J, Skarina, T, Mesa, N, Endres, M, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-15 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Putative oxidoreductase from Escherichia coli str. K-12
to be published
|
|
8EY3
 
 | | Contact-dependent growth inhibition (CDI) immunity protein from E. coli O32:H37 | | Descriptor: | Cys_rich_CPCC domain-containing protein, FE (III) ION, SODIUM ION | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Goulding, C.W, Hayes, C.S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-10-26 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Contact-dependent growth inhibition (CDI) immunity protein from E. coli O32:H37
To Be Published
|
|
3E9A
 
 | | Crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, SULFATE ION | | Authors: | Nocek, B, Mulligan, R, Kwon, K, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-21 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from Vibrio cholerae O1
biovar eltor str. N16961
To be Published
|
|
6PUB
 
 | | Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae in the Complex with Crystal Violet | | Descriptor: | CHLORIDE ION, CRYSTAL VIOLET, Chloramphenicol acetyltransferase, ... | | Authors: | Kim, Y, Maltseva, N, Kuhn, M, Stam, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structure of the Type B Chloramphenicol Acetyltransferase from Vibrio cholerae in the Complex with Crystal Violet
To Be Published
|
|
6OSS
 
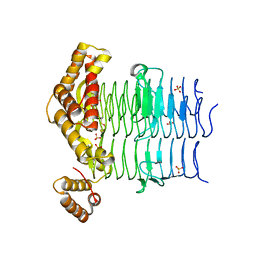 | | Crystal Structure of the Acyl-Carrier-Protein UDP-N-Acetylglucosamine O-Acyltransferase LpxA from Proteus mirabilis | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, PHOSPHITE ION, SULFATE ION | | Authors: | Kim, Y, Stogios, P, Skarina, T, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-29 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of the Acyl-Carrier-Protein UDP-N-Acetylglucosamine O-Acyltransferase LpxA from Proteus mirabilis
To Be Published
|
|
8UFM
 
 | | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2 | | Descriptor: | ACETATE ION, FORMIC ACID, Papain-like protease nsp3, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2
To Be Published
|
|
6OSU
 
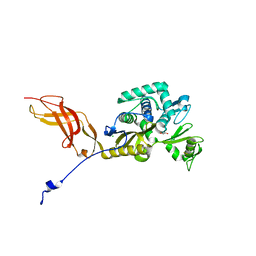 | | Crystal Structure of the D-alanyl-D-alanine carboxypeptidase DacD from Francisella tularensis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D-alanyl-D-alanine carboxypeptidase (Penicillin binding protein) family protein | | Authors: | Kim, Y, Stogios, P, Skarina, T, Di, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structure of the D-alanyl-D-alanine carboxypeptidase DacD from Francisella tularensis
To Be Published
|
|
4X3Z
 
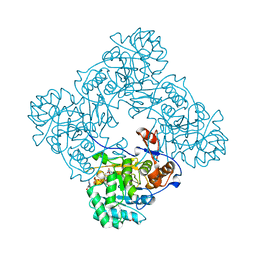 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with XMP and NAD | | Descriptor: | GLYCEROL, Inosine-5'-monophosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Osipiuk, J, MALTSEVA, N, KIM, Y, Mulligan, R, MAKOWSKA-GRZYSKA, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with XMP and NAD
to be published
|
|
6PFN
 
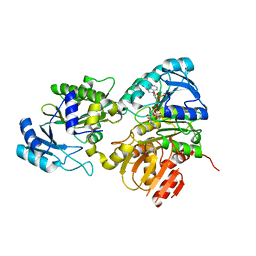 | | Succinyl-CoA synthase from Francisella tularensis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, COENZYME A, ... | | Authors: | Osipiuk, J, Maltseva, N, Jedrzejczak, R, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-06-21 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Succinyl-CoA synthase from Francisella tularensis
to be published
|
|
4X9O
 
 | | Beta-ketoacyl-ACP synthase III -2 (FabH2) (C113A) from Vibrio Cholerae soaked with octanoyl-CoA: conformational changes without clearly bound substrate | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2 | | Authors: | Hou, J, Cooper, D.R, Shabalin, I.G, Grabowski, M, Shumilin, I, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and enzymatic studies of beta-ketoacyl-(acyl carrier protein) synthase III (FabH) from Vibrio cholerae
to be published
|
|
6PO4
 
 | | 2.1 Angstrom Resolution Crystal Structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (mtnN) from Haemophilus influenzae PittII. | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, ... | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-03 | | Release date: | 2019-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase (mtnN) from Haemophilus influenzae PittII.
To Be Published
|
|
6OSX
 
 | | Crystal structure of uncharacterized protein ECL_02694 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Protein YmbA, ... | | Authors: | Chang, C, Evdokimova, E, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of uncharacterized protein ECL_02694
To Be Published
|
|
6OTV
 
 | | Crystal structure of putative isomerase EC2056 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Chang, C, Evdokimova, E, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-03 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of putative isomerase EC2056
To Be Published
|
|
8UFL
 
 | | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus | | Descriptor: | CHLORIDE ION, Papain-like protease nsp3, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus
To Be Published
|
|
4XCW
 
 | |
6P6P
 
 | |
