8J02
 
 | | Human KCNQ2(F104A)-CaM-PIP2-CBD complex in state II | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-04-09 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
8J01
 
 | | Human KCNQ2-CaM in complex with CBD and PIP2 | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-04-09 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
8YWY
 
 | | Crystal structure of SARS-Cov-2 main protease E166N mutant in complex with Bofutrelvir | | Descriptor: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhou, X.L, Wang, W.W, Zhang, J, Li, J. | | Deposit date: | 2024-04-01 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
8YKJ
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with X77 | | Descriptor: | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide, Non-structural protein 11 | | Authors: | Zhou, X.L, Lin, C, Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2024-03-05 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of coronaviral main proteases in complex with the non-covalent inhibitor X77.
Int.J.Biol.Macromol., 276, 2024
|
|
8YKL
 
 | | Crystal structure of MERS main protease in complex with X77 | | Descriptor: | N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide, ORF1a | | Authors: | Zhou, X.L, Lin, C, Zhang, J, Li, J. | | Deposit date: | 2024-03-05 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of coronaviral main proteases in complex with the non-covalent inhibitor X77.
Int.J.Biol.Macromol., 276, 2024
|
|
8YKK
 
 | | Crystal structure of SARS main protease in complex with X77 | | Descriptor: | 3C-like proteinase nsp5, N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-1H-imidazole-4-carboxamide | | Authors: | Zhou, X.L, Lin, C, Zhang, J, Li, J. | | Deposit date: | 2024-03-05 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of coronaviral main proteases in complex with the non-covalent inhibitor X77.
Int.J.Biol.Macromol., 276, 2024
|
|
8J03
 
 | |
7VVP
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2021-11-07 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
8Y63
 
 | | Cryo-EM structure of the C20:0 ceramide-bound FPR2-Gi complex | | Descriptor: | Cer(d18:0/20:0), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Jiang, C.T, Kong, W, Yu, X, Cai, K, Guo, L.L. | | Deposit date: | 2024-02-01 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Metabolic signaling of ceramides through the FPR2 receptor inhibits adipocyte thermogenesis.
Science, 388, 2025
|
|
8Y62
 
 | | Cryo-EM structure of the C16:0 ceramide-bound FPR2-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Sun, J.P, Jiang, C.T, Kong, W, Yu, X, Cai, K, Guo, L.L. | | Deposit date: | 2024-02-01 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Metabolic signaling of ceramides through the FPR2 receptor inhibits adipocyte thermogenesis.
Science, 388, 2025
|
|
8JWH
 
 | |
7WQH
 
 | | Crystal structure of HCoV-NL63 main protease with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhong, F.L, Zhou, X.L, Lin, C, Zeng, P, Li, J, Zhang, J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Basis of Main Proteases of Coronavirus Bound to Drug Candidate PF-07304814
J.Mol.Biol., 434, 2022
|
|
8K6Q
 
 | | Crystal structure of HOIL-1L LTM domain | | Descriptor: | RanBP-type and C3HC4-type zinc finger-containing protein 1 | | Authors: | Yan, Z, Pan, L.F. | | Deposit date: | 2023-07-25 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Mechanistic insights into the homo-dimerization of HOIL-1L and SHARPIN.
Biochem.Biophys.Res.Commun., 689, 2023
|
|
8K6P
 
 | | Crystal structure of SHARPIN LTM motif | | Descriptor: | Sharpin | | Authors: | Yan, Z, Pan, L.F. | | Deposit date: | 2023-07-25 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Mechanistic insights into the homo-dimerization of HOIL-1L and SHARPIN.
Biochem.Biophys.Res.Commun., 689, 2023
|
|
7YK1
 
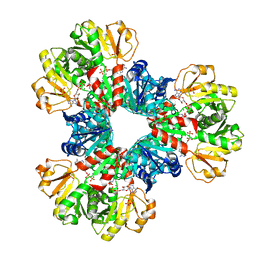 | | Structural basis of human PRPS2 filaments | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Lu, G.M, Hu, H.H, Liu, J.L. | | Deposit date: | 2022-07-21 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis of human PRPS2 filaments.
Cell Biosci, 13, 2023
|
|
7VFB
 
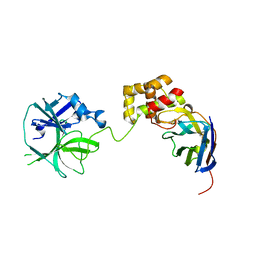 | | the complex of SARS-CoV2 3cl and NB2B4 | | Descriptor: | 3C-like proteinase, nb2b4 | | Authors: | Geng, Y, Sun, Z.C, Wang, L. | | Deposit date: | 2021-09-11 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An extended conformation of SARS-CoV-2 main protease reveals allosteric targets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VFA
 
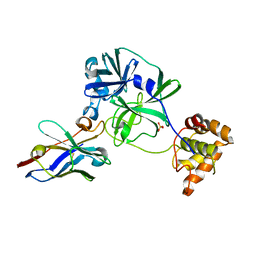 | | the complex of SARS-CoV2 3CL and NB1A2 | | Descriptor: | 3C-like proteinase, NB1A2, SULFATE ION | | Authors: | Sun, Z.C, Wang, L, Geng, Y. | | Deposit date: | 2021-09-11 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An extended conformation of SARS-CoV-2 main protease reveals allosteric targets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8X8A
 
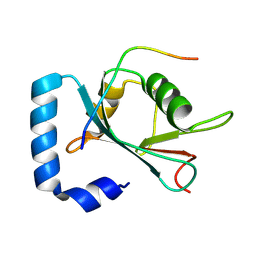 | | Crystal structure of STBD1 LIR motif in complex with GABARAPL1 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, Starch-binding domain-containing protein 1 | | Authors: | Zhang, Y.C, Pan, L.F. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Decoding the molecular mechanism of selective autophagy of glycogen mediated by autophagy receptor STBD1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8X8K
 
 | | Crystal structure of STBD1 CBM20 domain in complex with maltotetraose | | Descriptor: | GLYCEROL, Starch-binding domain-containing protein 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhang, Y.C, Pan, L.F. | | Deposit date: | 2023-11-27 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Decoding the molecular mechanism of selective autophagy of glycogen mediated by autophagy receptor STBD1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8WUR
 
 | | Crystal structure of SARS-Cov-2 main protease D48N mutant in complex with shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase nsp5 | | Authors: | Zhao, Z.Y, Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2023-10-21 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Basis for the Inhibition of SARS-CoV-2 M pro D48N Mutant by Shikonin and PF-07321332.
Viruses, 16, 2023
|
|
8UTE
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 27 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S)-6,6-difluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-31 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
6A4C
 
 | | Solution structure of MXAN_0049 | | Descriptor: | Uncharacterized protein MXAN_0049 | | Authors: | Chen, C, Feng, Y. | | Deposit date: | 2018-06-19 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of MXAN_0049
To Be Published
|
|
7CVQ
 
 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB2/YC3 and FT CORE1 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-3 and Zinc finger protein CONSTANS, FT CORE1 DNA forward strand, FT CORE1 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
7CVO
 
 | | crystal structure of Arabidopsis CO CCT domain in complex with NF-YB3/YC4 and FT CORE2 DNA | | Descriptor: | Chimera of Nuclear transcription factor Y subunit C-4 and Zinc finger protein CONSTANS, FT CORE2 DNA forward strand, FT CORE2 DNA reverse strand, ... | | Authors: | Lv, X, Du, J. | | Deposit date: | 2020-08-26 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex.
Plant Cell, 33, 2021
|
|
7V6Y
 
 | | Cryo-EM structure of Patched in lipid nanodisc - the wildtype, 3.5 angstrom (re-processed with dataset of 7dzq) | | Descriptor: | (2S)-2-azanyl-3-[[(2S)-3-butanoyloxy-2-dec-9-enoyloxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Luo, Y, Zhao, Y, Qu, Q, Li, D. | | Deposit date: | 2021-08-20 | | Release date: | 2021-09-22 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM study of patched in lipid nanodisc suggests a structural basis for its clustering in caveolae.
Structure, 29, 2021
|
|
