9D4V
 
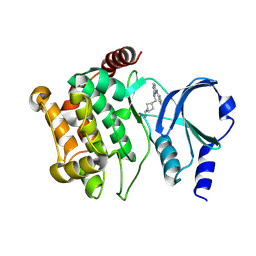 | | Structure of PAK1 in complex with compound 7 | | Descriptor: | N~2~-{[(1s,4s)-4-aminocyclohexyl]methyl}-N~4~-(5-cyclopropyl-1,3-thiazol-2-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase PAK 1 | | Authors: | Dementiev, A, Boone, C.D, Suto, R.K, Olland, A.M. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Identification of a p21-activated kinase 1 (PAK1) inhibitor with 10-fold selectivity against PAK2.
Bioorg.Med.Chem.Lett., 2025
|
|
9D4Y
 
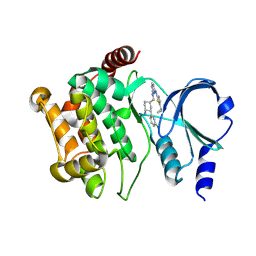 | | Structure of PAK1 in complex with compound 31 | | Descriptor: | N~2~-{[(1R,3R,4S)-4-amino-3-(3-chlorophenyl)cyclohexyl]methyl}-N~4~-(5-cyclopropyl-1,3-thiazol-2-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase PAK 1 | | Authors: | Fontano, E, Suto, R.K, Olland, A.M. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Identification of a p21-activated kinase 1 (PAK1) inhibitor with 10-fold selectivity against PAK2.
Bioorg.Med.Chem.Lett., 2025
|
|
9D53
 
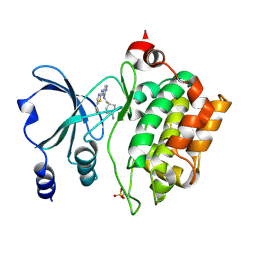 | | PAK4 in complex with compound 7 | | Descriptor: | N~2~-{[(1s,4s)-4-aminocyclohexyl]methyl}-N~4~-(5-cyclopropyl-1,3-thiazol-2-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase PAK 4 | | Authors: | Boone, C.D, Olland, A.M, Suto, R.K. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.468 Å) | | Cite: | Identification of a p21-activated kinase 1 (PAK1) inhibitor with 10-fold selectivity against PAK2.
Bioorg.Med.Chem.Lett., 2025
|
|
9D50
 
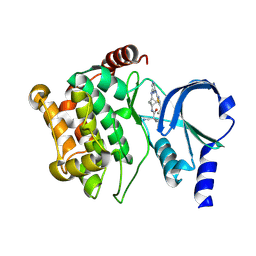 | | Structure of PAK1 in complex with compound 24 | | Descriptor: | 3-cyano-N-[3-({6-[(5-cyclopropyl-1,3-thiazol-2-yl)amino]pyrazin-2-yl}amino)bicyclo[1.1.1]pentan-1-yl]azetidine-3-carboxamide, Serine/threonine-protein kinase PAK 1 | | Authors: | Fontano, E, Suto, R.K, Olland, A.M. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Identification of a p21-activated kinase 1 (PAK1) inhibitor with 10-fold selectivity against PAK2.
Bioorg.Med.Chem.Lett., 2025
|
|
9D51
 
 | | Structure of PAK2 in complex with compound 12 | | Descriptor: | N~2~-{[(1s,4s)-4-aminocyclohexyl]methyl}-N~4~-[5-(trifluoromethyl)-1,3-thiazol-2-yl]pyrimidine-2,4-diamine, PAK-2p34 | | Authors: | Cakici, O, Suto, R.K, Olland, A.M. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of a p21-activated kinase 1 (PAK1) inhibitor with 10-fold selectivity against PAK2.
Bioorg.Med.Chem.Lett., 2025
|
|
9D4X
 
 | | Structure of PAK1 in complex with compound 16 | | Descriptor: | N~2~-{[(1R,3R,4S)-4-amino-3-(3-chlorophenyl)cyclohexyl]methyl}-N~4~-(5-cyclopropyl-1,3-thiazol-2-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase PAK 1 | | Authors: | Dementiev, A, Suto, R.K, Olland, A.M. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Identification of a p21-activated kinase 1 (PAK1) inhibitor with 10-fold selectivity against PAK2.
Bioorg.Med.Chem.Lett., 2025
|
|
4HOG
 
 | | Candida glabrata dihydrofolate reductase complexed with NADPH and 5-[3-(2-methoxy-4-phenylphenyl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine (UCP111H) | | Descriptor: | 5-[3-(2-methoxy-4-phenylphenyl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, CHLORIDE ION, Dihydrofolate Reductase, ... | | Authors: | Paulsen, J.L, Anderson, A.C. | | Deposit date: | 2012-10-22 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Propargyl-Linked Antifolates are Dual Inhibitors of Candida albicans and Candida glabrata.
J.Med.Chem., 57, 2014
|
|
4HOF
 
 | | Candida albicans dihydrofolate reductase complexed with NADPH and 5-[3-(2-methoxy-4-phenylphenyl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine (UCP111H) | | Descriptor: | 5-[3-(2-methoxy-4-phenylphenyl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Paulsen, J.L, Anderson, A.C. | | Deposit date: | 2012-10-22 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Propargyl-Linked Antifolates are Dual Inhibitors of Candida albicans and Candida glabrata.
J.Med.Chem., 57, 2014
|
|
4HOE
 
 | | Candida albicans dihydrofolate reductase complexed with NADPH and 5-[3-(2,5-dimethoxy-4-phenylphenyl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine (UCP111E) | | Descriptor: | 5-[3-(2,5-dimethoxy-4-phenylphenyl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, Dihydrofolate reductase, GLYCEROL, ... | | Authors: | Paulsen, J.L, Anderson, A.C. | | Deposit date: | 2012-10-22 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Propargyl-Linked Antifolates are Dual Inhibitors of Candida albicans and Candida glabrata.
J.Med.Chem., 57, 2014
|
|
8UWZ
 
 | | The structure of Raamsizumab in complex with VEGF121 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform VEGF121 of Vascular endothelial growth factor A, long form, ... | | Authors: | Singer, A.U, Bruce, H, Blazer, L, Sicheri, F, Adams, J.J, Sidhu, S.S. | | Deposit date: | 2023-11-08 | | Release date: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Raamsizumab: A Potent Synthetic Human Anti-VEGF Neutralizing Fab for Treatment of Ocular Neovascularization Diseases
to be published
|
|
8WCO
 
 | | (S)-citramalyl-CoA lyase | | Descriptor: | ACETYL COENZYME *A, Probable acyl-CoA lyase beta chain | | Authors: | Huang, Q, Bao, R. | | Deposit date: | 2023-09-13 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterization of itaconyl-CoA hydratase and citramalyl-CoA lyase involved in itaconate metabolism of Pseudomonas aeruginosa.
Structure, 32, 2024
|
|
8K5S
 
 | | The structure of EntE with 3-(prop-2-yn-1-yloxy)benzoic acid sulfamoyl adenosine | | Descriptor: | Enterobactin synthase component E, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-(3-prop-2-ynoxyphenyl)carbonylsulfamate | | Authors: | Miyanaga, A, Ishikawa, F. | | Deposit date: | 2023-07-24 | | Release date: | 2024-07-31 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biosynthetic Incorporation of Non-native Aryl Acid Building Blocks into Peptide Products Using Engineered Adenylation Domains.
Acs Chem.Biol., 19, 2024
|
|
8K5T
 
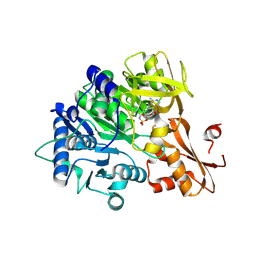 | | The structure of EntE with2-methyl-3-chloro-benzoic acid sulfamoyl adenosine | | Descriptor: | Enterobactin synthase component E, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-(3-chloranyl-2-methyl-phenyl)carbonylsulfamate | | Authors: | Miyanaga, A, Ishikawa, F. | | Deposit date: | 2023-07-24 | | Release date: | 2024-07-31 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biosynthetic Incorporation of Non-native Aryl Acid Building Blocks into Peptide Products Using Engineered Adenylation Domains.
Acs Chem.Biol., 19, 2024
|
|
8XLA
 
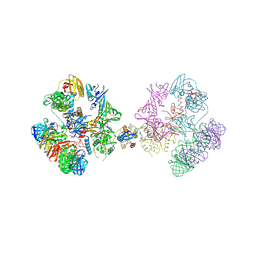 | | Mismatch Repair Complex | | Descriptor: | Beta sliding clamp, DNA mismatch repair protein MutL | | Authors: | Nirwal, S, Nair, D.T. | | Deposit date: | 2023-12-25 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of the MutL-CTD:processivity-clamp complex provides insight regarding strand discrimination in non-methyl-directed DNA mismatch repair.
Nucleic Acids Res., 53, 2025
|
|
1J3H
 
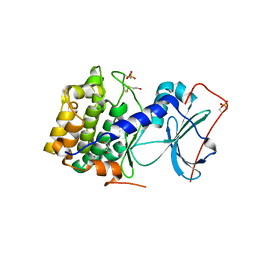 | | Crystal structure of apoenzyme cAMP-dependent protein kinase catalytic subunit | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Akamine, P, Madhusudan, Wu, J, Xuong, N.H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 2003-01-31 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dynamic Features of cAMP-dependent Protein Kinase Revealed by Apoenzyme Crystal Structure
J.Mol.Biol., 327, 2003
|
|
6HE0
 
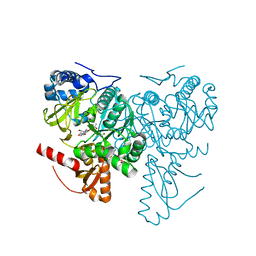 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in complex with 2-HIB-AMP and CoA in the thioesterfication state | | Descriptor: | 2-hydroxyisobutyryl-CoA synthetase, ADENOSINE MONOPHOSPHATE, COENZYME A, ... | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
6HDY
 
 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in the postadenylation state in complex with S3-HB-AMP | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, 2-hydroxyisobutyryl-CoA synthetase, SULFATE ION, ... | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
8KHG
 
 | | Itaconyl-CoA hydratase PaIch | | Descriptor: | CITRIC ACID, Itaconyl-CoA hydratase | | Authors: | Huang, Q, Bao, R. | | Deposit date: | 2023-08-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional characterization of itaconyl-CoA hydratase and citramalyl-CoA lyase involved in itaconate metabolism of Pseudomonas aeruginosa.
Structure, 32, 2024
|
|
8KHL
 
 | | (S)-citramalyl-CoA lyase | | Descriptor: | COENZYME A, Probable acyl-CoA lyase beta chain | | Authors: | Huang, Q, Bao, R. | | Deposit date: | 2023-08-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of itaconyl-CoA hydratase and citramalyl-CoA lyase involved in itaconate metabolism of Pseudomonas aeruginosa.
Structure, 32, 2024
|
|
6HE2
 
 | | Crystal structure of an open conformation of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in complex with 2-HIB-AMP and CoA | | Descriptor: | 2-hydroxyisobutyryl-CoA synthetase, ADENOSINE MONOPHOSPHATE, COENZYME A, ... | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
6HDX
 
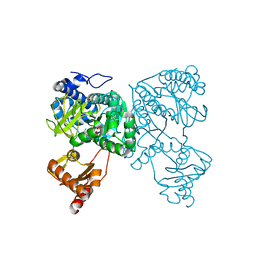 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in the postadenylation state in complex with R3-HIB-AMP | | Descriptor: | (2R)-3-HYDROXY-2-METHYLPROPANOIC ACID, 2-hydroxyisobutyryl-CoA synthetase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] (2~{R})-2-methyl-3-oxidanyl-propanoate | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
3ZIW
 
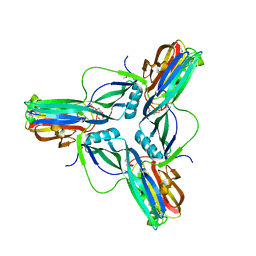 | | Clostridium perfringens enterotoxin, D48A mutation and N-terminal 37 residues deleted | | Descriptor: | HEAT-LABILE ENTEROTOXIN B CHAIN, HEXAETHYLENE GLYCOL | | Authors: | Yelland, T, Naylor, C.E, Savva, C.G, Basak, A.K. | | Deposit date: | 2013-01-14 | | Release date: | 2014-01-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a C. Perfringens Enterotoxin Mutant in Complex with a Modified Claudin-2 Extracellular Loop 2
J.Mol.Biol., 426, 2014
|
|
4KEB
 
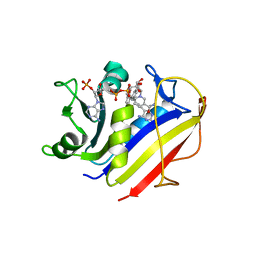 | | Human dihydrofolate reductase complexed with NADPH and 5-{3-[3-methoxy-5-(isoquin-5-yl)phenyl]but-1-yn-1-yl}6-ethylpyrimidine-2,4-diamine | | Descriptor: | 6-ethyl-5-{(3S)-3-[3-(isoquinolin-5-yl)-5-methoxyphenyl]but-1-yn-1-yl}pyrimidine-2,4-diamine, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | Lamb, K.M, Anderson, A.C. | | Deposit date: | 2013-04-25 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Elucidating features that drive the design of selective antifolates using crystal structures of human dihydrofolate reductase.
Biochemistry, 52, 2013
|
|
3ZIX
 
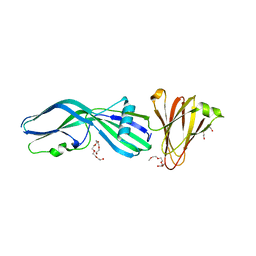 | | Clostridium perfringens Enterotoxin with the N-terminal 37 residues deleted | | Descriptor: | HEAT-LABILE ENTEROTOXIN B CHAIN, HEXAETHYLENE GLYCOL | | Authors: | Yelland, T, Naylor, C.E, Savva, C.G, Basak, A.K. | | Deposit date: | 2013-01-14 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a C. Perfringens Enterotoxin Mutant in Complex with a Modified Claudin-2 Extracellular Loop 2
J.Mol.Biol., 426, 2014
|
|
4KAK
 
 | | Crystal structure of human dihydrofolate reductase complexed with NADPH and 6-ethyl-5-[(3S)-3-[3-methoxy-5-(pyridine-4-yl)phenyl]but-1-yn-1-yl]pyrimidine-2,4-diamine (UCP1006) | | Descriptor: | 6-ethyl-5-{(3R)-3-[3-methoxy-5-(pyridin-4-yl)phenyl]but-1-yn-1-yl}pyrimidine-2,4-diamine, CALCIUM ION, Dihydrofolate reductase, ... | | Authors: | Lamb, K.M, Anderson, A.C. | | Deposit date: | 2013-04-22 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Elucidating features that drive the design of selective antifolates using crystal structures of human dihydrofolate reductase.
Biochemistry, 52, 2013
|
|
