4RLJ
 
 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium tuberculosis | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadA, (3R)-hydroxyacyl-ACP dehydratase subunit HadB, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Li, J, Dong, Y, Rao, Z.H. | | Deposit date: | 2014-10-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular basis for the inhibition of beta-hydroxyacyl-ACP dehydratase HadAB complex from Mycobacterium tuberculosis by flavonoid inhibitors.
Protein Cell, 6, 2015
|
|
4RLT
 
 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium tuberculosis complexed with Fisetin | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadA, (3R)-hydroxyacyl-ACP dehydratase subunit HadB, 3,7,3',4'-TETRAHYDROXYFLAVONE, ... | | Authors: | Li, J, Dong, Y, Rao, Z.H. | | Deposit date: | 2014-10-18 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Molecular basis for the inhibition of beta-hydroxyacyl-ACP dehydratase HadAB complex from Mycobacterium tuberculosis by flavonoid inhibitors.
Protein Cell, 6, 2015
|
|
9EQG
 
 | | CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with GABA and puerarin | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kasaragod, V.B, Aricescu, A.R. | | Deposit date: | 2024-03-21 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A brain-to-gut signal controls intestinal fat absorption.
Nature, 634, 2024
|
|
6KZ6
 
 | | Crystal structure of ASFV dUTPase | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R, MAGNESIUM ION | | Authors: | Guo, Y, Chen, C, Li, G.B, Cao, L, Wang, C.W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural Insight into African Swine Fever Virus dUTPase Reveals a Novel Folding Pattern in the dUTPase Family.
J.Virol., 94, 2020
|
|
1VFQ
 
 | |
6J18
 
 | | ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ESX-5 secretion system protein EccC5, MAGNESIUM ION | | Authors: | Wang, S.H, Li, J, Rao, Z.H. | | Deposit date: | 2018-12-28 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into substrate recognition by the type VII secretion system.
Protein Cell, 11, 2020
|
|
6J19
 
 | | ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ESAT-6-like protein EsxB, ESX-1 secretion system protein EccCb1, ... | | Authors: | Wang, S.H, Li, J, Rao, Z.H. | | Deposit date: | 2018-12-28 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Structural insights into substrate recognition by the type VII secretion system.
Protein Cell, 11, 2020
|
|
6JD5
 
 | | ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ESX conserved component EccC2. ESX-2 type VII secretion system protein. Possible membrane protein, MAGNESIUM ION | | Authors: | Wang, S.H, Li, J, Rao, Z.H. | | Deposit date: | 2019-01-31 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into substrate recognition by the type VII secretion system.
Protein Cell, 11, 2020
|
|
6J17
 
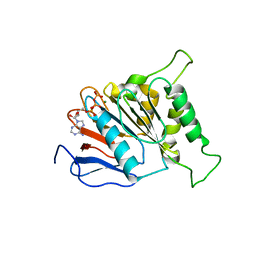 | | ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ESX-3 secretion system protein EccC3, MAGNESIUM ION | | Authors: | Wang, S.H, Li, J, Rao, Z.H. | | Deposit date: | 2018-12-28 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Structural insights into substrate recognition by the type VII secretion system.
Protein Cell, 11, 2020
|
|
6J72
 
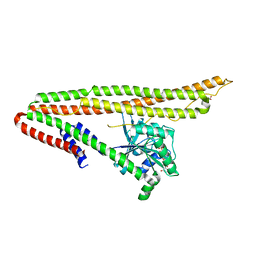 | | Crystal structure of IniA from Mycobacterium smegmatis with GTP bound | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Isoniazid inducible gene protein IniA, L(+)-TARTARIC ACID, ... | | Authors: | Wang, M.F, Guo, X.Y, Hu, J.J, Li, J, Rao, Z.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mycobacterial dynamin-like protein IniA mediates membrane fission.
Nat Commun, 10, 2019
|
|
6JD4
 
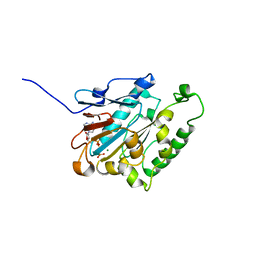 | | ATPase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ESX-1 secretion system protein EccCb1, MAGNESIUM ION | | Authors: | Wang, S.H, Li, J, Rao, Z.H. | | Deposit date: | 2019-01-31 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into substrate recognition by the type VII secretion system.
Protein Cell, 11, 2020
|
|
6J73
 
 | | Crystal structure of IniA from Mycobacterium smegmatis | | Descriptor: | Isoniazid inducible gene protein IniA | | Authors: | Wang, M.F, Guo, X.Y, Hu, J.J, Li, J, Rao, Z.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Mycobacterial dynamin-like protein IniA mediates membrane fission.
Nat Commun, 10, 2019
|
|
3DJU
 
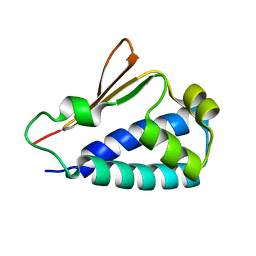 | | Crystal structure of human BTG2 | | Descriptor: | Protein BTG2 | | Authors: | Yang, X, Morita, M, Wang, H, Suzuki, T, Bartlam, M, Yamamoto, T. | | Deposit date: | 2008-06-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity
Nucleic Acids Res., 36, 2008
|
|
2K7X
 
 | |
3DJN
 
 | | Crystal structure of mouse TIS21 | | Descriptor: | Protein BTG2 | | Authors: | Yang, X, Morita, M, Wang, H, Suzuki, T, Bartlam, M, Yamamoto, T. | | Deposit date: | 2008-06-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity
Nucleic Acids Res., 36, 2008
|
|
3EB7
 
 | | Crystal Structure of Insecticidal Delta-Endotoxin Cry8Ea1 from Bacillus Thuringiensis at 2.2 Angstroms Resolution | | Descriptor: | ACETATE ION, Insecticidal Delta-Endotoxin Cry8Ea1, SULFATE ION | | Authors: | Guo, S, Ye, S, Song, F, Zhang, J, Wei, L, Shu, C.L. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Bacillus thuringiensis Cry8Ea1: An insecticidal toxin toxic to underground pests, the larvae of Holotrichia parallela.
J.Struct.Biol., 168, 2009
|
|
3FG2
 
 | |
5CA9
 
 | |
5CA8
 
 | |
8J8K
 
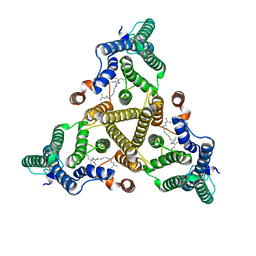 | | Membrane bound PRTase, C3 symmetry, acceptor bound | | Descriptor: | Decaprenyl-phosphate phosphoribosyltransferase, MONO-TRANS, OCTA-CIS DECAPRENYL-PHOSPHATE | | Authors: | Wu, F.Y, Gao, S, Zhang, L, Rao, Z.H. | | Deposit date: | 2023-05-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural analysis of phosphoribosyltransferase-mediated cell wall precursor synthesis in Mycobacterium tuberculosis.
Nat Microbiol, 9, 2024
|
|
8J8J
 
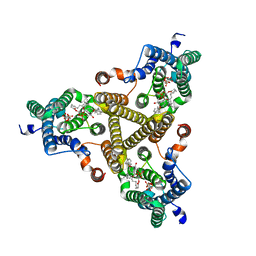 | | Membrane bound PRTase, C3 symmetry, donor bound | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Decaprenyl-phosphate phosphoribosyltransferase, ... | | Authors: | Wu, F.Y, Gao, S, Zhang, L, Rao, Z.H. | | Deposit date: | 2023-05-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural analysis of phosphoribosyltransferase-mediated cell wall precursor synthesis in Mycobacterium tuberculosis.
Nat Microbiol, 9, 2024
|
|
9KR5
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 3 | | Descriptor: | (6~{E})-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-[5-[(3~{S})-oxolan-3-yl]oxypyridin-3-yl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazinane-2,4-dione, 3C-like proteinase nsp5 | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-27 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSK
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 10 | | Descriptor: | 3C-like proteinase, 4-[4-chloranyl-2-[[(6E)-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-(5-methylpyridin-3-yl)-2,4-bis(oxidanylidene)-1,3,5-triazinan-1-yl]methyl]-5-fluoranyl-phenoxy]-2-fluoranyl-benzenecarbonitrile | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSI
 
 | | Crystal Structure of SARS-CoV-2 main protease in complex with compound 5 | | Descriptor: | (6E)-1-[[5-chloranyl-4-fluoranyl-2-(4-fluoranylphenoxy)phenyl]methyl]-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-(5-methoxypyridin-3-yl)-1,3,5-triazinane-2,4-dione, 3C-like proteinase nsp5 | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSH
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 1 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-pyridin-3-yl-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
