7Y65
 
 | | Cryo-EM structure of C5a peptide-bound C5aR1 in complex with Gi protein | | Descriptor: | C5a anaphylatoxin chemotactic receptor 1, C5apep peptide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
7Y66
 
 | | Cryo-EM structure of BM213-bound C5aR1 in complex with Gi protein | | Descriptor: | BM213 peptide, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
7Y64
 
 | | Cryo-EM structure of C5a-bound C5aR1 in complex with Gi protein | | Descriptor: | C5a anaphylatoxin, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
7KSI
 
 | |
7KSK
 
 | |
7KSJ
 
 | |
7WIN
 
 | |
4OLS
 
 | | The amidase-2 domain of LysGH15 | | Descriptor: | Endolysin, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Gu, J, Ouyang, S, Liu, Z.J, Han, W. | | Deposit date: | 2014-01-24 | | Release date: | 2014-06-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and biochemical characterization reveals LysGH15 as an unprecedented "EF-hand-like" calcium-binding phage lysin.
Plos Pathog., 10, 2014
|
|
4OLK
 
 | | The CHAP domain of LysGH15 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Endolysin | | Authors: | Gu, J, Ouyang, S, Liu, Z.J, Han, W. | | Deposit date: | 2014-01-24 | | Release date: | 2014-05-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Structural and biochemical characterization reveals LysGH15 as an unprecedented "EF-hand-like" calcium-binding phage lysin.
Plos Pathog., 10, 2014
|
|
3KPX
 
 | | Crystal Structure Analysis of photoprotein clytin | | Descriptor: | Apophotoprotein clytin-3, C2-HYDROPEROXY-COELENTERAZINE, CALCIUM ION | | Authors: | Titushin, M.S, Li, Y, Stepanyuk, G.A, Wang, B.-C, Lee, J, Vysotski, E.S, Liu, Z.-J. | | Deposit date: | 2009-11-17 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | NMR derived topology of a GFP-photoprotein energy transfer complex
J.Biol.Chem., 285, 2010
|
|
3UDC
 
 | | Crystal structure of a membrane protein | | Descriptor: | Small-conductance mechanosensitive channel, C-terminal peptide from Small-conductance mechanosensitive channel | | Authors: | Li, W, Ge, J, Yang, M. | | Deposit date: | 2011-10-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.355 Å) | | Cite: | Structure and molecular mechanism of an anion-selective mechanosensitive channel of small conductance
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5E9A
 
 | |
4OIP
 
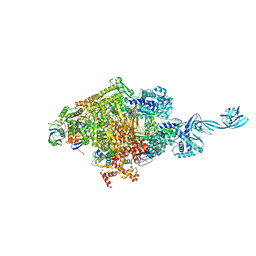 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077, ATP, and CMPcPP | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIR
 
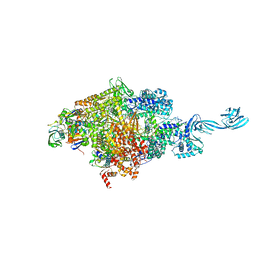 | | Crystal structure of Thermus thermophilus RNA polymerase transcription initiation complex soaked with GE23077 and rifamycin SV | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OIQ
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 and rifampicin | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.624 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
8H97
 
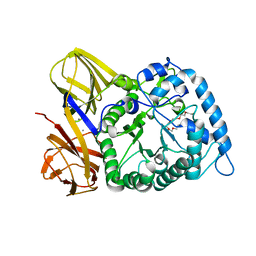 | | GH86 agarase Aga86A_Wa | | Descriptor: | Beta-agarase, CALCIUM ION, HEXAETHYLENE GLYCOL | | Authors: | Zhang, Y.Y, Dong, S, Feng, Y.G, Chang, Y.G. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural characterization on a beta-agarase Aga86A_Wa from Wenyingzhuangia aestuarii reveals the prevalent methyl-galactose accommodation capacity of GH86 enzymes at subsite -1.
Carbohydr Polym, 306, 2023
|
|
8HML
 
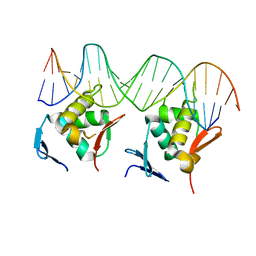 | |
6L87
 
 | |
7CGW
 
 | | Complex structure of PD-1 and tislelizumab Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of tislelizumab Fab, Light chain of tislelizumab Fab, ... | | Authors: | Hong, Y, Feng, Y.C, Liu, Y. | | Deposit date: | 2020-07-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Tislelizumab uniquely binds to the CC' loop of PD-1 with slow-dissociated rate and complete PD-L1 blockage.
Febs Open Bio, 11, 2021
|
|
3UDD
 
 | | Tankyrase-1 in complex with small molecule inhibitor | | Descriptor: | 3-(4-methoxyphenyl)-5-({[4-(4-methoxyphenyl)-5-methyl-4H-1,2,4-triazol-3-yl]sulfanyl}methyl)-1,2,4-oxadiazole, GLYCEROL, SULFATE ION, ... | | Authors: | Kirby, C.A, Stams, T. | | Deposit date: | 2011-10-28 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | [1,2,4]triazol-3-ylsulfanylmethyl)-3-phenyl-[1,2,4]oxadiazoles: antagonists of the Wnt pathway that inhibit tankyrases 1 and 2 via novel adenosine pocket binding.
J.Med.Chem., 55, 2012
|
|
3LFM
 
 | |
7CMW
 
 | | Complex structure of PARP1 catalytic domain with pamiparib | | Descriptor: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 1 | | Authors: | Feng, Y.C, Peng, H, Hong, Y, Liu, Y. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Pamiparib (BGB-290), a Potent and Selective Poly (ADP-ribose) Polymerase (PARP) Inhibitor in Clinical Development.
J.Med.Chem., 63, 2020
|
|
8HN6
 
 | | Crystal structure of monoclonal antibody complexed with SARS-CoV-2 RBD | | Descriptor: | Heavy chain of monoclonal antibody 3G10, Light chain of monoclonal antibody 3G10, Spike protein S1 | | Authors: | Qi, J, Chen, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Characterization of RBD-specific cross-neutralizing antibodies responses against SARS-CoV-2 variants from COVID-19 convalescents.
Front Immunol, 14, 2023
|
|
8HDJ
 
 | |
8HN7
 
 | | Crystal structure of monoclonal antibody complexed with SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of monoclonal antibody 3C11, Light chain of monoclonal antibody 3C11, ... | | Authors: | Qi, J, Chen, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-05-17 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Characterization of RBD-specific cross-neutralizing antibodies responses against SARS-CoV-2 variants from COVID-19 convalescents.
Front Immunol, 14, 2023
|
|
