4S24
 
 | | 1.7 Angstrom Crystal Structure of of Putative Modulator of Drug Activity (apo- form) from Yersinia pestis CO92 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Modulator of drug activity B, PENTAETHYLENE GLYCOL, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-01-19 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Crystal Structure of of Putative Modulator of Drug Activity (apo- form) from Yersinia pestis CO92.
TO BE PUBLISHED
|
|
4NHD
 
 | | Crystal structure of beta-ketoacyl-ACP synthase III (FabH) from Vibrio Cholerae in complex with Coenzyme A | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 1, CALCIUM ION, COENZYME A, ... | | Authors: | Hou, J, Zheng, H, Langner, K, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-04 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of beta-ketoacyl-ACP synthase III (FabH) from Vibrio Cholerae in complex with Coenzyme A
To be Published
|
|
4R01
 
 | | Crystal structure of SP1627, a putative NADH-flavin reductase, from Streptococcus pneumoniae TIGR4 | | Descriptor: | CHLORIDE ION, SULFATE ION, putative NADH-flavin reductase | | Authors: | Stogios, P.J, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of SP1627, a putative NADH-flavin reductase, from Streptococcus pneumoniae TIGR4
TO BE PUBLISHED
|
|
2L3M
 
 | | Solution structure of the putative copper-ion-binding protein from Bacillus anthracis str. Ames | | Descriptor: | Copper-ion-binding protein | | Authors: | Zhang, Y, Winsor, J, Dubrovska, I, Anderson, W, Radhakrishnan, I, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-16 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | To be published
To be Published
|
|
4RS2
 
 | | 1.55 Angstrom Crystal Structure of GNAT Family N-acetyltransferase (YhbS) from Escherichia coli in Complex with CoA | | Descriptor: | COENZYME A, Predicted acyltransferase with acyl-CoA N-acyltransferase domain | | Authors: | Minasov, G, Wawrzak, Z, Kuhn, M, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-11-06 | | Release date: | 2014-11-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 Angstrom Crystal Structure of GNAT Family N-acetyltransferase (YhbS) from Escherichia coli in Complex with CoA.
TO BE PUBLISHED
|
|
3HJV
 
 | | 1.7 Angstrom resolution crystal structure of an acyl carrier protein S-malonyltransferase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, Malonyl Coa-acyl carrier protein transacylase, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Anderson, S, Skarina, T, Onopriyenko, O, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom resolution crystal structure of an acyl carrier protein S-malonyltransferase from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
3I3O
 
 | | 2.06 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor' in complex with NAD-acetone | | Descriptor: | CACODYLATE ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Halavaty, A.S, Minasov, G, Skarina, T, Onopriyenko, O, Peterson, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | 2.06 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor' in complex with NAD-acetone
To be Published
|
|
3IMF
 
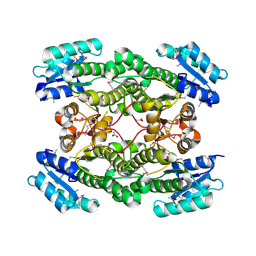 | | 1.99 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | ACETATE ION, Short chain dehydrogenase | | Authors: | Halavaty, A.S, Minasov, G, Skarina, T, Onopriyenko, O, Gordon, E, Peterson, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-10 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | 1.99 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
3E9A
 
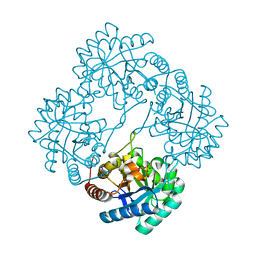 | | Crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, SULFATE ION | | Authors: | Nocek, B, Mulligan, R, Kwon, K, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-21 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from Vibrio cholerae O1
biovar eltor str. N16961
To be Published
|
|
4OC9
 
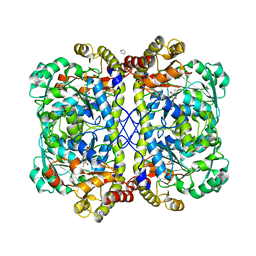 | | 2.35 Angstrom resolution crystal structure of putative O-acetylhomoserine (thiol)-lyase (metY) from Campylobacter jejuni subsp. jejuni NCTC 11168 with N'-Pyridoxyl-Lysine-5'-Monophosphate at position 205 | | Descriptor: | GLYCEROL, IMIDAZOLE, PHOSPHATE ION, ... | | Authors: | Halavaty, A.S, Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-12 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution crystal structure of putative O-acetylhomoserine (thiol)-lyase (metY) from Campylobacter jejuni subsp. jejuni NCTC 11168 with N'-Pyridoxyl-Lysine-5'-Monophosphate at position 205
To be Published
|
|
5SWU
 
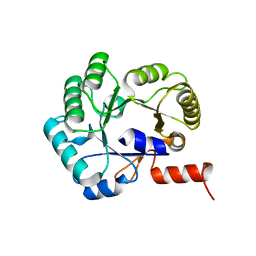 | |
3IJR
 
 | | 2.05 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor' in complex with NAD+ | | Descriptor: | MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Oxidoreductase, ... | | Authors: | Halavaty, A.S, Minasov, G, Skarina, T, Onopriyenko, O, Gordon, E, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor' in complex with NAD+
To be Published
|
|
5TKW
 
 | | 1.35 Angstrom Resolution Crystal Structure of a Pullulanase-specific Type II Secretion System Integral Cytoplasmic Membrane Protein GspL (N-terminal fragment; residues 1-237) from Klebsiella pneumoniae. | | Descriptor: | FORMIC ACID, Type II secretion system protein L | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-09 | | Release date: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | 1.35 Angstrom Resolution Crystal Structure of a Pullulanase-specific Type II Secretion System Integral Cytoplasmic Membrane Protein GspL (N-terminal fragment; residues 1-237) from Klebsiella pneumoniae.
To Be Published
|
|
3KHY
 
 | | Crystal Structure of a propionate kinase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | Propionate kinase | | Authors: | Brunzelle, J.S, Skarina, T, Sharma, S, Wang, Y, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-31 | | Release date: | 2010-01-19 | | Last modified: | 2014-10-01 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Crystal Structure of a propionate kinase from Francisella tularensis subsp. tularensis SCHU S4
To be Published
|
|
5TK2
 
 | | Crystal Structure of Uncharacterized Cupredoxin-like domain protein from Bacillus anthracis | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Cytochrome B, ... | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-06 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Uncharacterized Cupredoxin-like domain protein from
Bacillus anthracis
To Be Published
|
|
3KB8
 
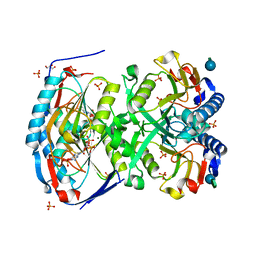 | | 2.09 Angstrom resolution structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-1) from Bacillus anthracis str. 'Ames Ancestor' in complex with GMP | | Descriptor: | GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE, Hypoxanthine phosphoribosyltransferase, ... | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-20 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | 2.09 Angstrom resolution structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-1) from Bacillus anthracis str. 'Ames Ancestor' in complex with GMP
To be Published
|
|
5U63
 
 | | Crystal structure of putative thioredoxin reductase from Haemophilus influenzae | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Michalska, K, Maltseva, N, Mulligan, R, Grimshaw, S, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of putative thioredoxin reductase from Haemophilus influenzae
To Be Published
|
|
3G25
 
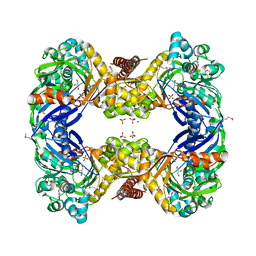 | | 1.9 Angstrom Crystal Structure of Glycerol Kinase (glpK) from Staphylococcus aureus in Complex with Glycerol. | | Descriptor: | GLYCEROL, Glycerol kinase, PHOSPHATE ION, ... | | Authors: | Minasov, G, Skarina, T, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of Glycerol Kinase (glpK) from Staphylococcus aureus in Complex with Glycerol.
TO BE PUBLISHED
|
|
5TSE
 
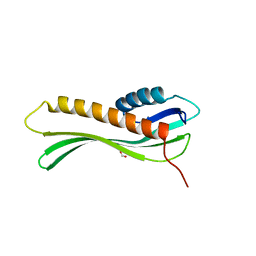 | | 2.35 Angstrom Crystal Structure Minor Lipoprotein from Acinetobacter baumannii. | | Descriptor: | FORMIC ACID, LPS-assembly lipoprotein LptE | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-28 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom Crystal Structure Minor Lipoprotein from Acinetobacter baumannii.
To Be Published
|
|
5TU0
 
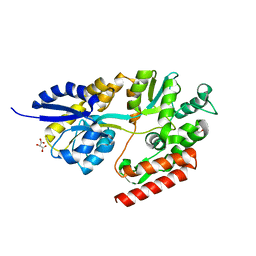 | | 1.9 Angstrom Resolution Crystal Structure of Maltose-Binding Periplasmic Protein MalE from Listeria monocytogenes in Complex with Maltose | | Descriptor: | Lmo2125 protein, TARTRONATE, TRIETHYLENE GLYCOL, ... | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-04 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of Maltose-Binding Periplasmic Protein MalE from Listeria monocytogenes in Complex with Maltose.
To Be Published
|
|
5TTA
 
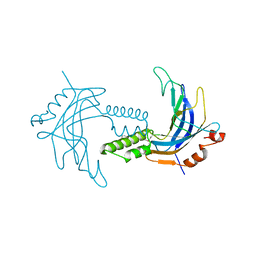 | | A 1.85A X-Ray Structure from Peptoclostridium difficile 630 of a Hypothetical Protein | | Descriptor: | Putative exported protein | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Cordona-Correa, A, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-02 | | Release date: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A 1.85A X-Ray Structure from Peptoclostridium difficile 630 of a Hypothetical Protein
To Be Published
|
|
3GIU
 
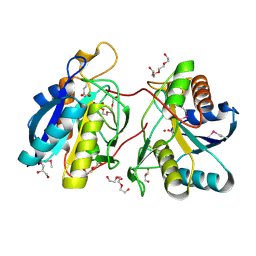 | | 1.25 Angstrom Crystal Structure of Pyrrolidone-Carboxylate Peptidase (pcp) from Staphylococcus aureus | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-06 | | Release date: | 2009-03-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25 Angstrom Crystal Structure of Pyrrolidone-Carboxylate Peptidase (pcp) from Staphylococcus aureus
TO BE PUBLISHED
|
|
5TV7
 
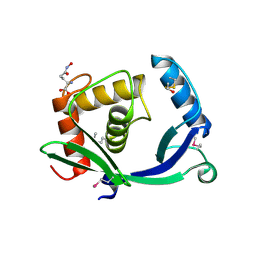 | | 2.05 Angstrom Resolution Crystal Structure of Peptidoglycan-Binding Protein from Clostridioides difficile in Complex with Glutamine Hydroxamate. | | Descriptor: | GLUTAMINE HYDROXAMATE, Putative peptidoglycan-binding/hydrolysing protein | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Winsor, J, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Resolution Crystal Structure of Peptidoglycan-Binding Protein from Clostridioides difficile in Complex with Glutamine Hydroxamate.
To Be Published
|
|
5UJS
 
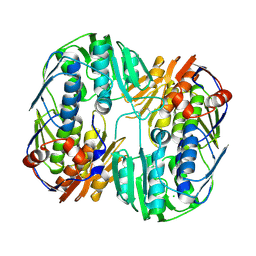 | | 2.45 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Campylobacter jejuni. | | Descriptor: | CHLORIDE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Stam, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | 2.45 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Campylobacter jejuni.
To Be Published
|
|
5UOU
 
 | | High resolution structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 | | Descriptor: | 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline (OHCU) decarboxylase | | Authors: | Chang, C, Li, H, Bearden, J, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-01 | | Release date: | 2017-02-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High resolution structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase from Klebsiella pneumoniae subsp. pneumoniae MGH 78578
To Be Published
|
|
