6X5N
 
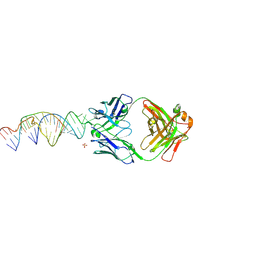 | | Crystal structure of a stabilized PAN ENE bimolecular triplex with a GC-clamped polyA tail, in complex with Fab-BL-3,6 | | Descriptor: | Heavy chain Fab Bl-3 6, Light chain Fab BL-3 6, SULFATE ION, ... | | Authors: | Swain, M, Li, M, Woldawer, A, LeGrice, S.F.J. | | Deposit date: | 2020-05-26 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dynamic bulge nucleotides in the KSHV PAN ENE triple helix provide a unique binding platform for small molecule ligands.
Nucleic Acids Res., 49, 2021
|
|
3FVH
 
 | |
3C5L
 
 | |
2K8F
 
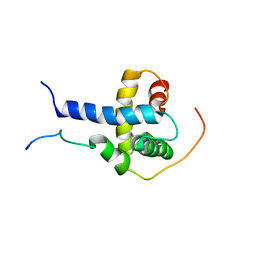 | | Structural Basis for the Regulation of p53 Function by p300 | | Descriptor: | Cellular tumor antigen p53, Histone acetyltransferase p300 | | Authors: | Bai, Y, Feng, H, Jenkins, L.M, Durell, S.R, Wiodawer, A, Appella, E. | | Deposit date: | 2008-09-08 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for p300 Taz2-p53 TAD1 Binding and Modulation by Phosphorylation.
Structure, 17, 2009
|
|
3BX9
 
 | |
4PTI
 
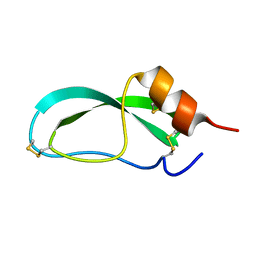 | | THE GEOMETRY OF THE REACTIVE SITE AND OF THE PEPTIDE GROUPS IN TRYPSIN, TRYPSINOGEN AND ITS COMPLEXES WITH INHIBITORS | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Huber, R, Kukla, D, Ruehlmann, A, Epp, O, Formanek, H, Deisenhofer, J, Steigemann, W. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors
Acta Crystallogr.,Sect.B, 39, 1983
|
|
3BXB
 
 | |
3BXC
 
 | |
3BXA
 
 | |
2OGR
 
 | |
4EDS
 
 | |
4EDO
 
 | |
4HE4
 
 | |
3U8A
 
 | |
3U8C
 
 | |
3EE1
 
 | |
2ICR
 
 | | Red fluorescent protein zRFP574 from Zoanthus sp. | | Descriptor: | Red fluorescent protein zoanRFP, SULFATE ION | | Authors: | Pletnev, S, Pletneva, N, Tikhonova, T, Pletnev, V. | | Deposit date: | 2006-09-13 | | Release date: | 2007-10-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Refined crystal structures of red and green fluorescent proteins from the button polyp Zoanthus.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3IXO
 
 | |
2PXW
 
 | | Crystal Structure of N66D Mutant of Green Fluorescent Protein from Zoanthus sp. at 2.4 A Resolution (Transition State) | | Descriptor: | GFP-like fluorescent chromoprotein FP506 | | Authors: | Pletnev, S.V, Pletneva, N.V, Tikhonova, T.V, Pletnev, V.Z. | | Deposit date: | 2007-05-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Refined crystal structures of red and green fluorescent proteins from the button polyp Zoanthus.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2OJK
 
 | | Crystal Structure of Green Fluorescent Protein from Zoanthus sp at 2.2 A Resolution | | Descriptor: | GFP-like fluorescent chromoprotein FP506 | | Authors: | Pletneva, N.V, Pletnev, S.V, Tikhonova, T.V, Pletnev, V.Z. | | Deposit date: | 2007-01-12 | | Release date: | 2007-09-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined crystal structures of red and green fluorescent proteins from the button polyp Zoanthus.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2PXS
 
 | | Crystal Structure of N66D Mutant of Green Fluorescent Protein from Zoanthus sp. at 2.2 A Resolution (Mature State) | | Descriptor: | GFP-like fluorescent chromoprotein FP506 | | Authors: | Pletnev, S.V, Pletneva, N.V, Tikhonova, T.V, Pletnev, V.Z. | | Deposit date: | 2007-05-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined crystal structures of red and green fluorescent proteins from the button polyp Zoanthus.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4JEO
 
 | |
3GB3
 
 | |
3GL4
 
 | |
4JGE
 
 | |
