2ZVR
 
 | |
3AU5
 
 | |
3WYB
 
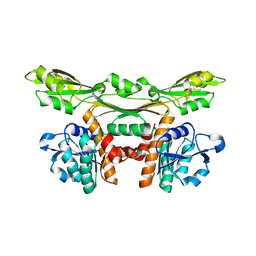 | | Structure of a meso-diaminopimelate dehydrogenase | | Descriptor: | Meso-diaminopimelate D-dehydrogenase | | Authors: | Sakuraba, H, Akita, H, Ohshima, T. | | Deposit date: | 2014-08-25 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into the thermostable NADP(+)-dependent meso-diaminopimelate dehydrogenase from Ureibacillus thermosphaericus
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3WYC
 
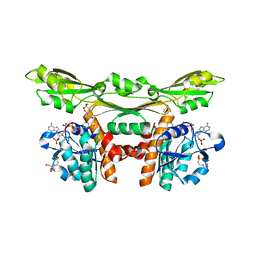 | | Structure of a meso-diaminopimelate dehydrogenase in complex with NADP | | Descriptor: | 2-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-ETHANESULFONIC ACID, Meso-diaminopimelate D-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sakuraba, H, Akita, H, Ohshima, T. | | Deposit date: | 2014-08-25 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural insight into the thermostable NADP(+)-dependent meso-diaminopimelate dehydrogenase from Ureibacillus thermosphaericus
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3W31
 
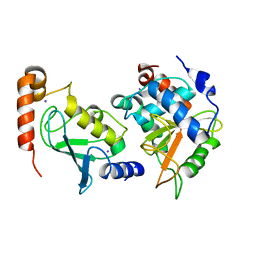 | | Structual basis for the recognition of Ubc13 by the Shigella flexneri effector OspI | | Descriptor: | IODIDE ION, ORF169b, Ubiquitin-conjugating enzyme E2 N | | Authors: | Nishide, A, Kim, M, Takagi, K, Sasakawa, C, Mizushima, T. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structural basis for the recognition of Ubc13 by the Shigella flexneri effector OspI.
J.Mol.Biol., 425, 2013
|
|
3W6Z
 
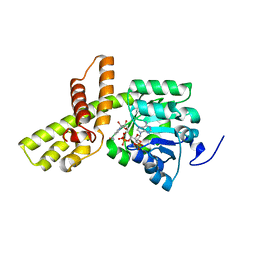 | |
3W6U
 
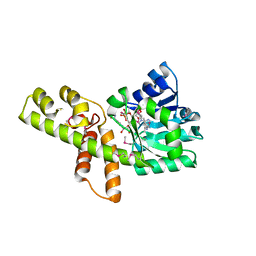 | |
1WNU
 
 | | Structure of Archaeal Trans-Editing Protein AlaX in complex with L-serine | | Descriptor: | SERINE, ZINC ION, alanyl-tRNA synthetase | | Authors: | Sokabe, M, Okada, A, Nakashima, T, Yao, M, Tanaka, I. | | Deposit date: | 2004-08-09 | | Release date: | 2005-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of alanine discrimination in editing site
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1X5L
 
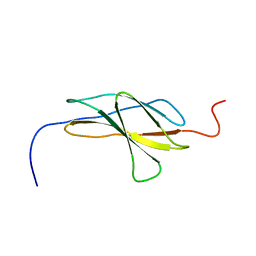 | |
1WPL
 
 | | Crystal structure of the inhibitory form of rat GTP cyclohydrolase I/GFRP complex | | Descriptor: | 7,8-DIHYDROBIOPTERIN, GTP cyclohydrolase I, GTP cyclohydrolase I feedback regulatory protein, ... | | Authors: | Maita, N, Hatakeyama, K, Okada, K, Hakoshima, T. | | Deposit date: | 2004-09-08 | | Release date: | 2004-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of biopterin-induced inhibition of GTP cyclohydrolase I by GFRP, its feedback regulatory protein
J.Biol.Chem., 279, 2004
|
|
1X61
 
 | |
1WE0
 
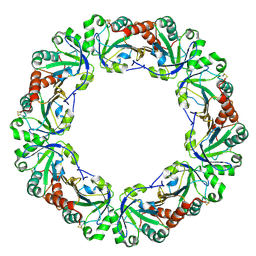 | | Crystal structure of peroxiredoxin (AhpC) from Amphibacillus xylanus | | Descriptor: | AMMONIUM ION, alkyl hydroperoxide reductase C | | Authors: | Kitano, K, Kita, A, Hakoshima, T, Niimura, Y, Miki, K. | | Deposit date: | 2004-05-21 | | Release date: | 2005-03-29 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of decameric peroxiredoxin (AhpC) from Amphibacillus xylanus
Proteins, 59, 2005
|
|
1X5Q
 
 | |
1WXO
 
 | | Structure of Archaeal Trans-Editing Protein AlaX in complex with zinc | | Descriptor: | ZINC ION, alanyl-tRNA synthetase | | Authors: | Sokabe, M, Okada, A, Nakashima, T, Yao, M, Tanaka, I. | | Deposit date: | 2005-01-27 | | Release date: | 2005-07-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Molecular basis of alanine discrimination in editing site
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1X5M
 
 | |
7F76
 
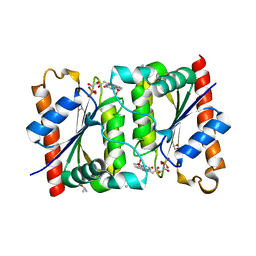 | |
1Y56
 
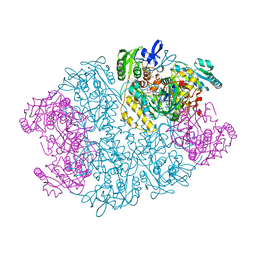 | | Crystal structure of L-proline dehydrogenase from P.horikoshii | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Tsuge, H, Kawakami, R, Sakuraba, H, Ago, H, Miyano, M, Aki, K, Katunuma, N, Ohshima, T. | | Deposit date: | 2004-12-02 | | Release date: | 2005-07-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Crystal structure of a novel FAD-, FMN-, and ATP-containing L-proline dehydrogenase complex from Pyrococcus horikoshii
J.Biol.Chem., 280, 2005
|
|
293D
 
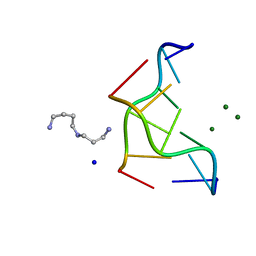 | | INTERACTION BETWEEN THE LEFT-HANDED Z-DNA AND POLYAMINE-2: THE CRYSTAL STRUCTURE OF THE D(CG)3 AND SPERMIDINE COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ohishi, H, Nakanishi, I, Inubushi, K, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J, Hakoshima, T, Tomita, K. | | Deposit date: | 1996-10-09 | | Release date: | 1996-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Interaction between the left-handed Z-DNA and polyamine-2. The crystal structure of the d(CG)3 and spermidine complex.
FEBS Lett., 391, 1996
|
|
292D
 
 | | INTERACTION BETWEEN THE LEFT-HANDED Z-DNA AND POLYAMINE:THE CRYSTAL STRUCTURE OF THE D(CG)3 AND N-(2-AMINOETHYL)-1,4-DIAMINOBUTANE COMPLEX | | Descriptor: | 1-(AMINOETHYL)AMINO-4-AMINOBUTANE, DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Ohishi, H, Kunisawa, S, Van Der Marel, G, Van Boom, J.H, Rich, A, Wang, A.H.-J, Tomita, K, Hakoshima, T. | | Deposit date: | 1991-10-09 | | Release date: | 1996-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Interaction between the left-handed Z-DNA and polyamine. The crystal structure of the d(CG)3 and N-(2-aminoethyl)-1,4-diamino-butane complex.
FEBS Lett., 284, 1991
|
|
1X63
 
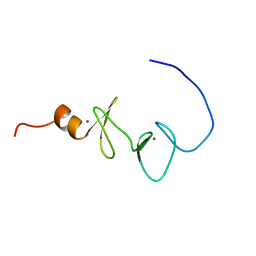 | |
1X62
 
 | |
2A25
 
 | | Crystal structure of Siah1 SBD bound to the peptide EKPAAVVAPITTG from SIP | | Descriptor: | Calcyclin-binding protein peptide, Ubiquitin ligase SIAH1, ZINC ION | | Authors: | Santelli, E, Leone, M, Li, C, Fukushima, T, Preece, N.E, Olson, A.J, Ely, K.R, Reed, J.C, Pellecchia, M, Liddington, R.C, Matsuzawa, S. | | Deposit date: | 2005-06-21 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Siah1-Siah-interacting Protein Interactions and Insights into the Assembly of an E3 Ligase Multiprotein Complex
J.Biol.Chem., 280, 2005
|
|
2A26
 
 | | Crystal structure of the N-terminal, dimerization domain of Siah Interacting Protein | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Calcyclin-binding protein, SULFATE ION | | Authors: | Santelli, E, Leone, M, Li, C, Fukushima, T, Preece, N.E, Olson, A.J, Ely, K.R, Reed, J.C, Pellecchia, M, Liddington, R.C, Matsuzawa, S. | | Deposit date: | 2005-06-21 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Analysis of Siah1-Siah-interacting Protein Interactions and Insights into the Assembly of an E3 Ligase Multiprotein Complex
J.Biol.Chem., 280, 2005
|
|
3VLD
 
 | | Crystal structure of yeast proteasome interacting protein | | Descriptor: | DNA mismatch repair protein HSM3 | | Authors: | Takagi, K, Kim, S, Kato, K, Tanaka, K, Saeki, Y, Mizushima, T. | | Deposit date: | 2011-12-01 | | Release date: | 2012-02-22 | | Last modified: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for specific recognition of Rpt1, an ATPase subunit of the 26S proteasome, by a proteasome-dedicated chaperone Hsm3
J.Biol.Chem., 287, 2012
|
|
3VLF
 
 | | Crystal structure of yeast proteasome interacting protein | | Descriptor: | 26S protease regulatory subunit 7 homolog, DNA mismatch repair protein HSM3 | | Authors: | Takagi, K, Kim, S, Kato, K, Tanaka, K, Saeki, Y, Mizushima, T. | | Deposit date: | 2011-12-01 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for specific recognition of Rpt1, an ATPase subunit of the 26S proteasome, by a proteasome-dedicated chaperone Hsm3
J.Biol.Chem., 287, 2012
|
|
