3J6C
 
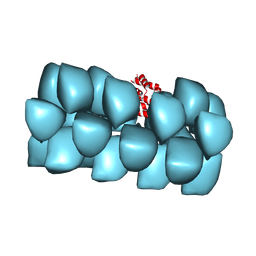 | | Cryo-EM structure of MAVS CARD filament | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | Xu, H, He, X, Zheng, H, Huang, L.J, Hou, F, Yu, Z, de la Cruz, M.J, Borkowski, B, Zhang, X, Chen, Z.J, Jiang, Q.-X. | | Deposit date: | 2014-02-04 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Structural basis for the prion-like MAVS filaments in antiviral innate immunity.
Elife, 3, 2014
|
|
2B36
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (InhA) inhibited by 5-pentyl-2-phenoxyphenol | | Descriptor: | 5-PENTYL-2-PHENOXYPHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sullivan, T.J, Truglio, J.J, Novichenok, P, Stratton, C, Zhang, X, Kaur, T, Johnson, F, Boyne, M.S, Amin, A. | | Deposit date: | 2005-09-19 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High Affinity InhA Inhibitors with Activity against Drug-Resistant Strains
of Mycobacterium tuberculosis
ACS Chem.Biol., 1, 2006
|
|
6D2K
 
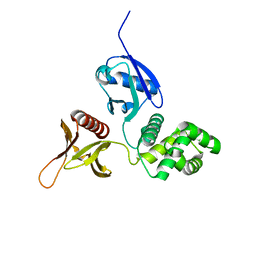 | | Crystal structure of the FERM domain of mouse FARP2 | | Descriptor: | FERM, ARHGEF and pleckstrin domain-containing protein 2 | | Authors: | He, X, Zhang, X. | | Deposit date: | 2018-04-13 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analyses of FERM domain-mediated membrane localization of FARP1.
Sci Rep, 8, 2018
|
|
6CIJ
 
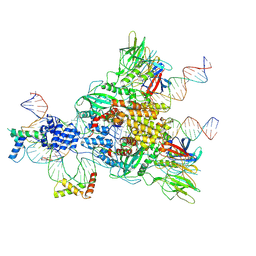 | | Cryo-EM structure of mouse RAG1/2 HFC complex containing partial HMGB1 linker(3.9 A) | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (41-MER), ... | | Authors: | Chen, X, Kim, M, Chuenchor, W, Cui, Y, Zhang, X, Zhou, Z.H, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
6CG0
 
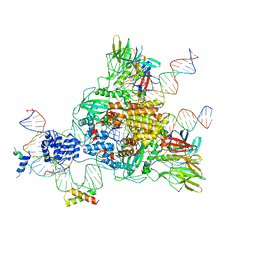 | | Cryo-EM structure of mouse RAG1/2 HFC complex (3.17 A) | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (41-MER), ... | | Authors: | Chen, X, Kim, M, Chuenchor, W, Cui, Y, Zhang, X, Zhou, Z.H, Gellert, M, Yang, W. | | Deposit date: | 2018-02-19 | | Release date: | 2018-04-25 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
2BJV
 
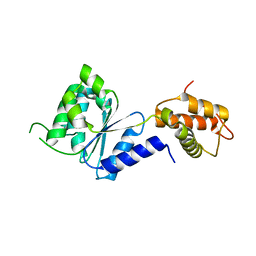 | | Crystal Structure of PspF(1-275) R168A mutant | | Descriptor: | PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Beuron, F, Niwa, H, Bordes, P, Wigneshweraraj, S, Keetch, C.A, Robinson, C.V, Buck, M, Zhang, X. | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights Into the Activity of Enhancer-Binding Proteins
Science, 307, 2005
|
|
2BJW
 
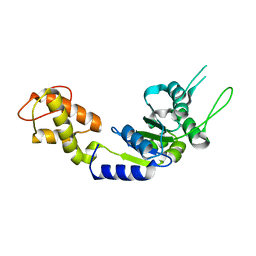 | | PspF AAA domain | | Descriptor: | PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Beuron, F, Niwa, H, Bordes, P, Wigneshweraraj, S, Keetch, C.A, Robinson, C.V, Buck, M, Zhang, X. | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights Into the Activity of Enhancer-Binding Proteins
Science, 307, 2005
|
|
5OQ4
 
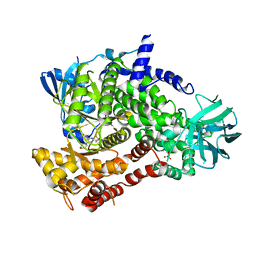 | | PQR309 - a Potent, Brain-Penetrant, Orally Bioavailable, pan-Class I PI3K/mTOR Inhibitor as Clinical Candidate in Oncology | | Descriptor: | 5-(4,6-dimorpholin-4-yl-1,3,5-triazin-2-yl)-4-(trifluoromethyl)pyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Williams, R.L, Zhang, X. | | Deposit date: | 2017-08-10 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 5-(4,6-Dimorpholino-1,3,5-triazin-2-yl)-4-(trifluoromethyl)pyridin-2-amine (PQR309), a Potent, Brain-Penetrant, Orally Bioavailable, Pan-Class I PI3K/mTOR Inhibitor as Clinical Candidate in Oncology.
J. Med. Chem., 60, 2017
|
|
2B35
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (InhA) inhibited by triclosan | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Sullivan, T.J, Truglio, J.J, Novichenok, P, Stratton, C, Zhang, X, Kaur, T, Johnson, F, Boyne, M.S, Amin, A. | | Deposit date: | 2005-09-19 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High Affinity InhA Inhibitors with Activity against Drug-Resistant Strains
of Mycobacterium tuberculosis
ACS Chem.Biol., 1, 2006
|
|
5OAF
 
 | | Human Rvb1/Rvb2 heterohexamer in INO80 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Aramayo, R.J, Bythell-Douglas, R, Ayala, R, Willhoft, O, Wigley, D, Zhang, X. | | Deposit date: | 2017-06-21 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Cryo-EM structures of the human INO80 chromatin-remodeling complex.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2B37
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (InhA) inhibited by 5-octyl-2-phenoxyphenol | | Descriptor: | 5-OCTYL-2-PHENOXYPHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sullivan, T.J, Truglio, J.J, Novichenok, P, Stratton, C, Zhang, X, Kaur, T, Johnson, F, Boyne, M.S, Amin, A. | | Deposit date: | 2005-09-19 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High Affinity InhA Inhibitors with Activity against Drug-Resistant Strains
of Mycobacterium tuberculosis
ACS Chem.Biol., 1, 2006
|
|
9OC4
 
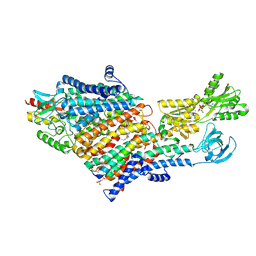 | | High-resolution cryo-EM structure of KdpFABC in the E1P-ADP state in lipid nanodisc | | Descriptor: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hussein, A.K, Zhang, X, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2025-04-23 | | Release date: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Conduction pathway for potassium through the E. coli pump KdpFABC
Biorxiv, 2025
|
|
6D2Q
 
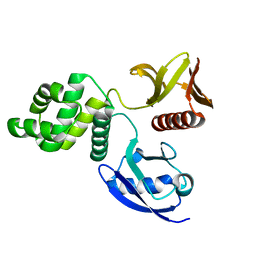 | | Crystal structure of the FERM domain of zebrafish FARP1 | | Descriptor: | FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) | | Authors: | Kuo, Y.C, Zhang, X. | | Deposit date: | 2018-04-13 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural analyses of FERM domain-mediated membrane localization of FARP1.
Sci Rep, 8, 2018
|
|
1RUN
 
 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP)/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA (5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*TP*CP*G )-3'), DNA (5'-D(*GP*CP*GP*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Parkinson, G.N, Gunasekera, A, Vojtechovsky, J, Zhang, X, Kunkel, T.A, Berman, H.M, Ebright, R.H. | | Deposit date: | 1996-05-26 | | Release date: | 1997-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aromatic hydrogen bond in sequence-specific protein DNA recognition.
Nat.Struct.Biol., 3, 1996
|
|
6D21
 
 | |
4QNR
 
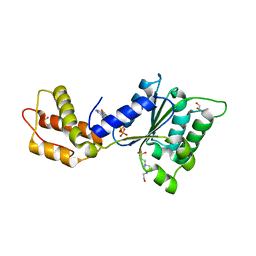 | | CRYSTAL STRUCTURE OF PSPF(1-265) E108Q MUTANT bound to ATP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
1R7R
 
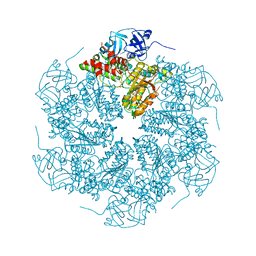 | | The crystal structure of murine p97/VCP at 3.6A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Huyton, T, Pye, V.E, Briggs, L.C, Flynn, T.C, Beuron, F, Kondo, H, Ma, J, Zhang, X, Freemont, P.S. | | Deposit date: | 2003-10-22 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The crystal structure of murine p97/VCP at 3.6A
J.Struct.Biol., 144, 2003
|
|
4QOS
 
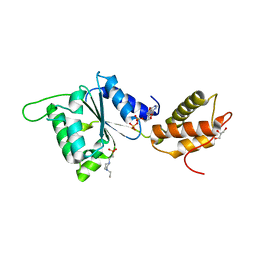 | | CRYSTAL STRUCTURE OF PSPF(1-265) E108Q MUTANT bound to ADP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-20 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
4QNM
 
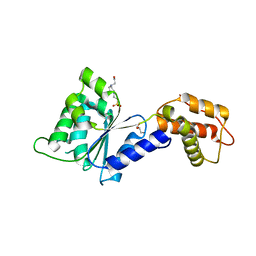 | | CRYSTAL STRUCTURE of PSPF(1-265) E108Q MUTANT | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Psp operon transcriptional activator | | Authors: | Darbari, V.C, Lawton, E, Lu, D, Burrows, P.C, Wiesler, S, Joly, N, Zhang, N, Zhang, X, Buck, M. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.628 Å) | | Cite: | Molecular basis of nucleotide-dependent substrate engagement and remodeling by an AAA+ activator.
Nucleic Acids Res., 42, 2014
|
|
1S3S
 
 | | Crystal structure of AAA ATPase p97/VCP ND1 in complex with p47 C | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase (TER ATPase) (15S Mg(2+)- ATPase p97 subunit) (Valosin containing protein) (VCP) [Contains: Valosin], p47 protein | | Authors: | Dreveny, I, Kondo, H, Uchiyama, K, Shaw, A, Zhang, X, Freemont, P.S. | | Deposit date: | 2004-01-14 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47.
Embo J., 23, 2004
|
|
7FAU
 
 | | Structure Determination of the NB1B11-RBD Complex | | Descriptor: | NB_1B11, Spike protein S1, ZINC ION | | Authors: | Shi, Z.Z, Li, X.X, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C, Cui, Q.Q, Qiao, H.R, Lan, Z.Y, Zhang, X. | | Deposit date: | 2021-07-07 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of nanobodies neutralizing SARS-CoV-2 variants.
Structure, 30, 2022
|
|
8R64
 
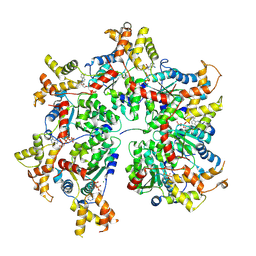 | | Cryo-EM structure of the FIGNL1 AAA hexamer bound to RAD51 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA repair protein RAD51 homolog 1, ... | | Authors: | Carver, A, Yates, L.A, Zhang, X. | | Deposit date: | 2023-11-20 | | Release date: | 2024-09-04 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of FIGNL1 in dissociating RAD51 from DNA and chromatin.
Science, 387, 2025
|
|
4EW0
 
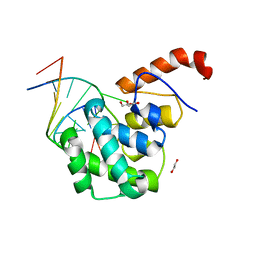 | | mouse MBD4 glycosylase domain in complex with a G:5hmU (5-hydroxymethyluracil) mismatch | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*GP*(5HU)P*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
6LBA
 
 | | Cryo-EM structure of the AtMLKL2 tetramer | | Descriptor: | Protein kinase family protein | | Authors: | Lisa, M, Huang, M, Zhang, X, Ryohei, T.N, Leila, B.K, Isabel, M.L.S, Florence, J, Viera, K, Dmitry, L, Jane, E.P, James, M.M, Kay, H, Paul, S.L, Chai, J, Takaki, M. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the AtMLKL3 tetramer
To Be Published
|
|
4EVV
 
 | | mouse MBD4 glycosylase domain in complex with a G:T mismatch | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*TP*GP*TP*GP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2012-04-26 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
